| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,142,903 – 6,143,017 |
| Length | 114 |
| Max. P | 0.596126 |

| Location | 6,142,903 – 6,143,017 |
|---|---|
| Length | 114 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.57 |
| Shannon entropy | 0.49843 |
| G+C content | 0.56253 |
| Mean single sequence MFE | -39.34 |
| Consensus MFE | -18.80 |
| Energy contribution | -18.29 |
| Covariance contribution | -0.51 |
| Combinations/Pair | 1.65 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.596126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6142903 114 - 22422827 ------UCACAUUUCGAUUACCACCAAGAAGGUGUGCACUCAAGAGCUGCCACUGCCCGAGGGCGUGGACGUCAUCCAUGCCUCUCCGGCCGCCGGUCAUCUGAGCAGCUCCAGCAUUUA ------...............((((.....))))(((......(((((((....(((.(((((((((((.....)))))))).))).)))...(((....))).)))))))..))).... ( -41.50, z-score = -2.21, R) >droSec1.super_4 5552176 114 + 6179234 ------GCACAUUUCGAUUACCACGAAGAAAGUGUGCACCCAAGAGCUGCCGCUGCCCGAGGGCGUGGACGUCAUCCAUGCCUCGCCGGCCGCCGGUCACCUGAGCAGCUCAAGCAUUUA ------((((((((((.......))))....))))))......(((((((.((.((((((((.((((((.....)))))))))))..))).))(((....))).)))))))......... ( -46.20, z-score = -3.00, R) >droYak2.chrX 2146789 114 - 21770863 ------GCACAUCUCGAUUACCACCAAGAAGGUGUGUACCCAAGAGCUGCCUCUGCCCGAGGGCGUGGACGUCAUCCAUGCCUCUCCGGCCGCCGGUCACCUGAGCAGCUCCAGCAUUUA ------((.......(..(((((((.....)))).)))..)..(((((((....(((.(((((((((((.....)))))))).))).)))...(((....))).)))))))..))..... ( -45.50, z-score = -3.15, R) >droEre2.scaffold_4690 15113004 114 - 18748788 ------GCACAUCUCGAUUACCACGAAGAAGGUGUGCACCCAAGAGCUGCCUCUGCCCGAGGGCGUGGACGUCAUCCAUGCCUCUCCGGCCGCAGGUCACCUGAGCAGCUCCAGCAUUUA ------(((((((((............).))))))))......(((((((....(((.(((((((((((.....)))))))).))).)))..((((...)))).)))))))......... ( -49.50, z-score = -3.76, R) >droAna3.scaffold_13417 805203 114 - 6960332 ------GCACAUUUCCAUCACCACCAAGAAGGUGUGCACCCAGGAGCUGCCCCUGCCCGAGGGCGUGGACGUGAUCCAUGCCUCCCCGGCUGCCGGCCACCUGAGCAGCUCCAGCAUCUA ------(((((...((.((........)).))))))).....((((((((........(..((((((((.....))))))))..)..(((.....)))......))))))))........ ( -44.80, z-score = -1.76, R) >dp4.chrXL_group1e 10309517 114 - 12523060 ------GCACAUCUCCAUAACCACGAAAAAGGUGUGCACGCAGGAGCUGCCGCUACCCGAGGGCGUGGAUGUGAUACAUGCCUCCCCGGCAGCGGGCCACUUGAGCAGCUCCAGCAUUUA ------((((((((...............))))))))..((.((((((((((((.((.(.(((((((.........)))))))..).)).))))(....)....)))))))).))..... ( -45.76, z-score = -2.07, R) >droPer1.super_69 132984 114 + 344178 ------GCACAUCUCAAUAACCACGAAAAAGGUGUGCACUCAGGAGCUGCCGCUACCCGAGGGCGUGGAUGUGAUACAUGCCUCCCCGGCAGCGGGCCACUUGAGCAGCUCCAGCAUUUA ------((((((((...............)))))))).....((((((((((((.((.(.(((((((.........)))))))..).)).))))(....)....))))))))........ ( -43.36, z-score = -1.80, R) >droWil1.scaffold_181150 2435606 114 + 4952429 ------ACAUAUAUCCAUUACUACCAAAAAGGUUUGCACACAAGAGCUACCACUGCCCGAGGGUGUGGAUGUUAUACAUGCCUCACCAGCAGCUGGCCAUUUGAGUAGCUCGAGCAUUUA ------................(((.....))).(((......(((((((((..(((.(..((((.(((((.....))).)).)))).....).)))....)).)))))))..))).... ( -29.30, z-score = -0.54, R) >droVir3.scaffold_13042 2350099 114 + 5191987 ------GCACAUCUCCAUCACCACGCAAAAGGUGUGCACCCAGGAGCUGCCAUUGCCGGAGGGCGUCGAUGUCAUCCAUGCCUCGCCGGCGGCGGGCCAUUUGAGCAGCUCGAGCAUUUA ------((((((((...............))))))))......(((((((((..(((((.((((((.(((...))).))))))..)))))((....))...)).)))))))......... ( -45.86, z-score = -1.35, R) >droMoj3.scaffold_6328 778008 114 - 4453435 ------GCACAUCUCGAUUACCACGAAGAAGGUCUGCACCCAGGAGCUGCCCCUGCCGGAGGGUGUGGAUGUCAUUCAUGCCUCCCCGGCGGCCGGGCACCUGAGCAGCUCGAGCAUUUA ------......(((((..(((........)))((((...((((...((((((((((((.((((((((((...))))))))))..)))))))..))))))))).)))).)))))...... ( -47.30, z-score = -1.51, R) >droGri2.scaffold_15081 1068504 114 + 4274704 ------ACACAUCUCGAUAACAACGAAGAAGGUGUGCACCCAGGAGCUACCGUUACCGGAGGGCGUCGAUGUCAUUCAUGCCUCACCGGCCGCUGGCCAUUUGAGCAGCUCGAGCAUUUA ------.((((((((............).))))))).......(((((.......((((.((((((.((......))))))))..))))..(((.(.....).))))))))......... ( -31.50, z-score = 0.59, R) >anoGam1.chr3L 37630546 114 + 41284009 ------GCACGUUAACAUUUCCACAACGAAAGUGUGCACCCAGGAGCUACCACUACCGGACGGCGUGGAUGUCAUCCAUGCUGCACCGGCAGCCGGUCAUCUGAGCUCGUCCUCGAUCUA ------(((((....).........((....))))))......((((((((.((.((((.((((((((((...))))))))))..)))).))..)))......)))))............ ( -37.50, z-score = -1.25, R) >apiMel3.GroupUn 380237063 120 + 399230636 AAAUAGUCAUGUUCUCAUUACUACUACAAAAGUAUGUACUCAAGAAUUACCAUUACCAGAUAGCGUUGAAGUAAUACAUGCAGCACCAGCAGCUGGACAUUUAAGCAGCUCUUCCAUUUA .(((.((...(((((...(((((((.....)))).)))....))))).)).)))....(..((.((((...(((....(.((((.......)))).)...)))..)))).))..)..... ( -15.60, z-score = 1.40, R) >triCas2.ChLG2 6135616 114 + 12900155 ------CCAUAUUAAUAUCACCGUGACGAGAGAAGUUAAACAAAUUUUGCCGAUUCCUGACGGUGUGGAGAUCGUUCAUGCUGCCCCAGCAGCUGGUCAUUUGAGUUCGGCGUCCAUUUA ------................(.(((....((((((.....))))))((((((((.((((...(((((.....)))))(((((....)))))..))))...))).)))))))))..... ( -27.10, z-score = -0.03, R) >consensus ______GCACAUCUCCAUUACCACGAAGAAGGUGUGCACCCAAGAGCUGCCACUGCCCGAGGGCGUGGAUGUCAUCCAUGCCUCCCCGGCAGCCGGCCACCUGAGCAGCUCCAGCAUUUA .......((((((.................)))))).......(((((((..........((((((...........))))))..((((...))))........)))))))......... (-18.80 = -18.29 + -0.51)

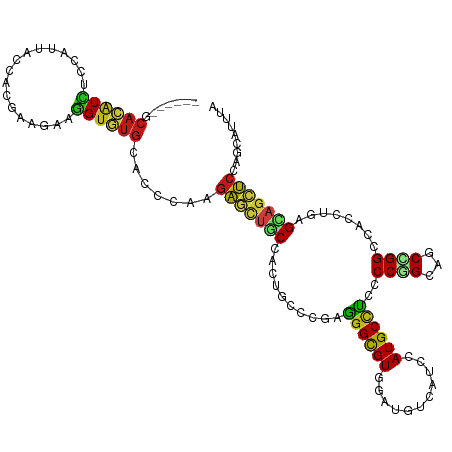
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:19:59 2011