| Sequence ID | dm3.chr2L |
|---|---|
| Location | 10,390,618 – 10,390,709 |
| Length | 91 |
| Max. P | 0.985010 |

| Location | 10,390,618 – 10,390,709 |
|---|---|
| Length | 91 |
| Sequences | 13 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 66.40 |
| Shannon entropy | 0.64414 |
| G+C content | 0.55991 |
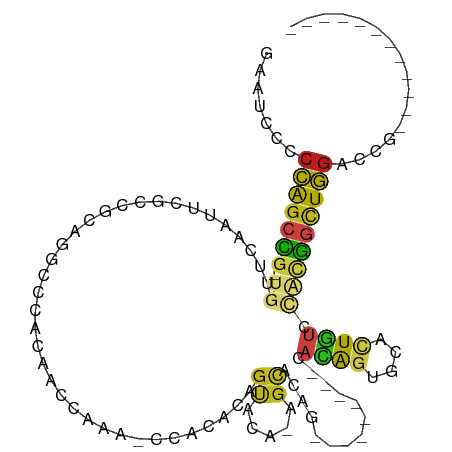
| Mean single sequence MFE | -25.44 |
| Consensus MFE | -9.74 |
| Energy contribution | -9.13 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.92 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.985010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
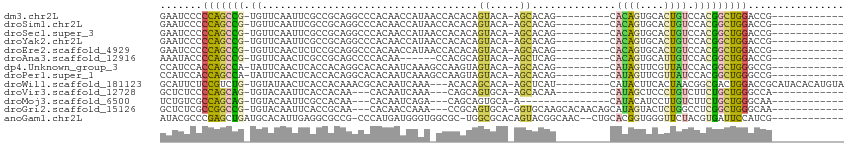
>dm3.chr2L 10390618 91 + 23011544 GAAUCCCCCAGCCG-UGUUCAAUUCGCCGCAGGCCCACAACCAUAACCACACAGUACA-AGCACAG---------CACAGUGCACUGUCCACGGCUGGACCG------------ .......(((((((-((........(((...)))................(((((...-.((((..---------....))))))))).)))))))))....------------ ( -28.90, z-score = -3.64, R) >droSim1.chr2L 10180387 91 + 22036055 GAAUCCCCCAGCCG-UGUUCAAUUCGCCGCAGGCCCACAACCAUAACCACACAGUACA-AGCACAG---------CACAGUGCACUGUCCACGGCUGGACCG------------ .......(((((((-((........(((...)))................(((((...-.((((..---------....))))))))).)))))))))....------------ ( -28.90, z-score = -3.64, R) >droSec1.super_3 5818529 91 + 7220098 GAAUCCCCCAGCCG-UGUUCAAUUCGCCGCAGGCCCACAACCAUAACCACACAGUACA-AGCACAG---------CACAGUGCACUGUCCACGGCUGGACCG------------ .......(((((((-((........(((...)))................(((((...-.((((..---------....))))))))).)))))))))....------------ ( -28.90, z-score = -3.64, R) >droYak2.chr2L 13071342 91 + 22324452 GAAUCCCCCAGCCG-UGUUCAAUUCGCCGCAGGCCCACAACCAUAACCACACAGUACA-AGCACAG---------CACAGUGCACUGUCCACGGCUGGACCG------------ .......(((((((-((........(((...)))................(((((...-.((((..---------....))))))))).)))))))))....------------ ( -28.90, z-score = -3.64, R) >droEre2.scaffold_4929 11000024 91 + 26641161 GAAUCCCCCAGCCG-UGUUCAACUCUCCGCAGGCCCACAACCAUAACCACACAGUACA-AGCACAG---------CACAGUGCACUGUCCACGGCUGGACCG------------ .......(((((((-((..............((.......))........(((((...-.((((..---------....))))))))).)))))))))....------------ ( -27.20, z-score = -3.25, R) >droAna3.scaffold_12916 14235953 85 + 16180835 AAAUACCCCAGCCG-UGUUCAACUCGCCGCAGCCCCACAA------CCACGCAGUACA-AGCUCAG---------CACAGUGCAUUGUCCACGGCUGGACCG------------ .......(((((((-((..(((..(((.((.((.......------....))(((...-.)))..)---------)...)))..)))..)))))))))....------------ ( -25.20, z-score = -2.55, R) >dp4.Unknown_group_3 71699 91 + 156060 CCAUCCACCAGCCA-UAUUCAACUCACCACAGGCACACAAUCAAAGCCAAGUAGUACA-AGCACAG---------CAUAGUUCGUUAUCCACGGCUGGGCCG------------ .......((((((.-................(((...........)))..((.(.((.-(((....---------....))).))...).))))))))....------------ ( -18.00, z-score = -2.32, R) >droPer1.super_1 1349397 91 + 10282868 CCAUCCACCAGCCA-UAUUCAACUCACCACAGGCACACAAUCAAAGCCAAGUAGUACA-AGCACAG---------CAUAGUUCGUUAUCCACGGCUGGGCCG------------ .......((((((.-................(((...........)))..((.(.((.-(((....---------....))).))...).))))))))....------------ ( -18.00, z-score = -2.32, R) >droWil1.scaffold_181123 1015442 100 + 1459946 GCAUUCUCCGUCUG-UGUAUAACUCACCACAAACGCACAAUCAAA---ACACAGCACA-AGCUCAU---------CAUACUUCACUAACGGCGACUGGACCGCAUACACAUGUA ............((-(((((......(((....(((.........---....(((...-.)))...---------.........(....))))..))).....))))))).... ( -12.90, z-score = 0.13, R) >droVir3.scaffold_12728 269640 85 - 808307 GCUCUCCCCAGCAG-UGUACAAUUCACCACAA---CACAAUCAAA---CAGCAGUGCA-AGCACAA---------CAUAGCUCCCUGUCUUCUGCUGGGCCA------------ ......((((((((-(((.............)---))).......---..((((.(..-(((....---------....))).)))))....)))))))...------------ ( -19.42, z-score = -1.30, R) >droMoj3.scaffold_6500 1684137 79 - 32352404 UCUGUCGCCAGCAG-UGUACAAUUCGCCACAA---CACAAUCAGA---CAGCAGUGCA-A---------------CAUACAUCCUUGUCUUCUGCUGGGCAA------------ ......(((....(-(((.............)---))).......---((((((....-(---------------((........)))...)))))))))..------------ ( -18.62, z-score = -0.56, R) >droGri2.scaffold_15126 7673348 94 + 8399593 GCUCUCGCCGGCCG-UGUACAAUUCACCGCAA---CACAACCAAA---CCGCAGUGCA-GGUGCAAGCACAACAGCAUAGUACUCUGGCCUCGGCUGGGCAA------------ ((((..((((((((-.((((....(((((((.---..(.......---..)...))).-))))...((......))...))))..)))))..))).))))..------------ ( -29.90, z-score = -0.55, R) >anoGam1.chr2L 35117724 98 + 48795086 AUACGCCCGAGCUGAUGCACAUUGAGGCGCCG-CCCAUGAUGGGUGGCGC-UGGCGCACAGUACGGCAAC--CUGCACGGUGGGUUCUACGUGAUUCCAUCG------------ ....(((.(.((((.(((.(.....(((((((-(((.....)))))))))-).).))))))).))))...--.....((((((..((.....))..))))))------------ ( -45.90, z-score = -2.53, R) >consensus GAAUCCCCCAGCCG_UGUUCAAUUCGCCGCAGGCCCACAACCAAA_CCACACAGUACA_AGCACAG_________CACAGUGCACUGUCCACGGCUGGACCG____________ .......(((((((.......................................((.....))..............((((....))))...)))))))................ ( -9.74 = -9.13 + -0.60)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:30:00 2011