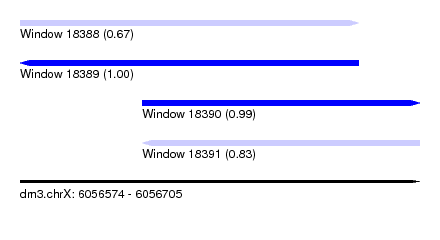
| Sequence ID | dm3.chrX |
|---|---|
| Location | 6,056,574 – 6,056,705 |
| Length | 131 |
| Max. P | 0.999220 |

| Location | 6,056,574 – 6,056,685 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 71.17 |
| Shannon entropy | 0.39664 |
| G+C content | 0.56059 |
| Mean single sequence MFE | -30.74 |
| Consensus MFE | -22.93 |
| Energy contribution | -22.93 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.672610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
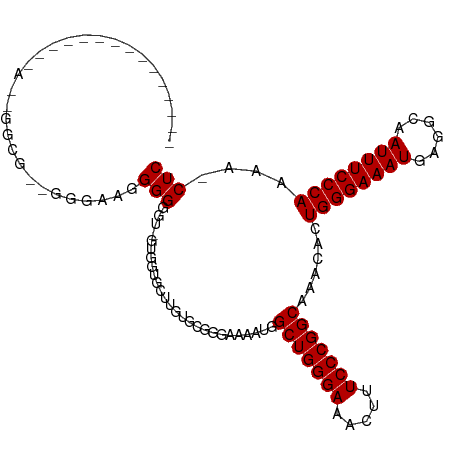
>dm3.chrX 6056574 111 + 22422827 CAAAGGGGGUGGGGGAAAGGCGGAGGGGAAGGGGGU-GUUGUGCUUGUGCGGGAAAAUGGCUGGGAAACUUUCCCGGCAAACACUGGGAAAUGAGGCAAUUUCCCAAAA-CUC ....(((((((..(.(.(((((.((.(........)-.)).))))).).).........(((((((.....)))))))...))))(((((((......)))))))....-))) ( -31.70, z-score = -0.47, R) >droEre2.scaffold_4690 15034814 87 + 18748788 --------------------------CAAGGGGGGCAGGGGGAGGGGGGCGGUAAAAUGGCUGGGAAACUUUCCCGGCAAACACUGGGAAAUGAGGCAAUUUCCCAAAAACUC --------------------------...(((..((.(((..((.....((((......)))).....))..))).))......((((((((......))))))))....))) ( -28.10, z-score = -1.72, R) >droSim1.chrX 4781680 100 + 17042790 ------------CAAAGGGGCGAGGGGAGGGGGCGUUGUGGUGCUUGUGCGGGAAAAUGGCUGGGAAACUUUCCCGGCAAACACUGGGAAAUGAGGCAAUUUCCCAAAA-CUC ------------....(((.....((((((..((.(..(((((.((((.((((((.......((....)))))))))))).)))).....)..).))..))))))....-))) ( -32.41, z-score = -1.04, R) >consensus _______________A__GGCG__GGGAAGGGGGGU_GUGGUGCUUGUGCGGGAAAAUGGCUGGGAAACUUUCCCGGCAAACACUGGGAAAUGAGGCAAUUUCCCAAAA_CUC ..............................(((..........................(((((((.....)))))))......((((((((......))))))))....))) (-22.93 = -22.93 + 0.00)

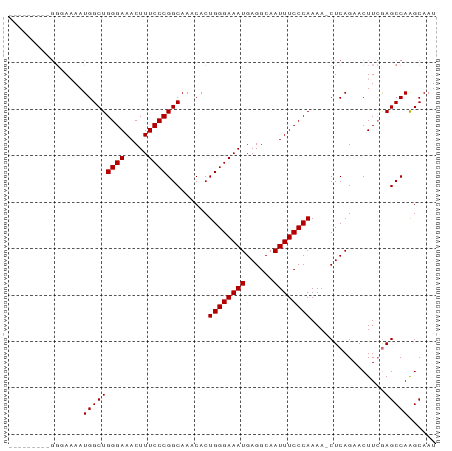
| Location | 6,056,574 – 6,056,685 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 71.17 |
| Shannon entropy | 0.39664 |
| G+C content | 0.56059 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -20.69 |
| Energy contribution | -22.13 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.43 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.72 |
| SVM RNA-class probability | 0.999220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
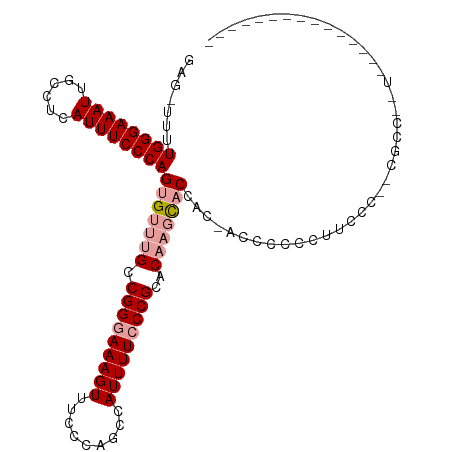
>dm3.chrX 6056574 111 - 22422827 GAG-UUUUGGGAAAUUGCCUCAUUUCCCAGUGUUUGCCGGGAAAGUUUCCCAGCCAUUUUCCCGCACAAGCACAAC-ACCCCCUUCCCCUCCGCCUUUCCCCCACCCCCUUUG (((-...((((((((......))))))))(((((((.(((((((((.........)))))))))..)))))))...-...........)))...................... ( -29.80, z-score = -4.78, R) >droEre2.scaffold_4690 15034814 87 - 18748788 GAGUUUUUGGGAAAUUGCCUCAUUUCCCAGUGUUUGCCGGGAAAGUUUCCCAGCCAUUUUACCGCCCCCCUCCCCCUGCCCCCCUUG-------------------------- (((...(((((((((......))))))))).....((.((((.....)))).))...............)))...............-------------------------- ( -18.70, z-score = -1.39, R) >droSim1.chrX 4781680 100 - 17042790 GAG-UUUUGGGAAAUUGCCUCAUUUCCCAGUGUUUGCCGGGAAAGUUUCCCAGCCAUUUUCCCGCACAAGCACCACAACGCCCCCUCCCCUCGCCCCUUUG------------ (((-...((((((((......))))))))(((((((.(((((((((.........)))))))))..)))))))................))).........------------ ( -29.80, z-score = -4.11, R) >consensus GAG_UUUUGGGAAAUUGCCUCAUUUCCCAGUGUUUGCCGGGAAAGUUUCCCAGCCAUUUUCCCGCACAAGCACCAC_ACCCCCCUUCCC__CGCC__U_______________ .......((((((((......))))))))(((((((.(((((((((.........)))))))))..)))))))........................................ (-20.69 = -22.13 + 1.45)

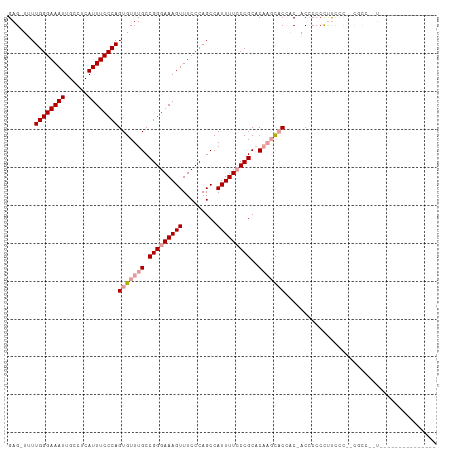
| Location | 6,056,614 – 6,056,705 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 88.38 |
| Shannon entropy | 0.17791 |
| G+C content | 0.46872 |
| Mean single sequence MFE | -30.40 |
| Consensus MFE | -23.27 |
| Energy contribution | -23.27 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.990692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6056614 91 + 22422827 UGCUUGUGCGGGAAAAUGGCUGGGAAACUUUCCCGGCAAACACUGGGAAAUGAGGCAAUUUCCCAAAA-CUCAGAACUUCGAGCCAAGCAAU ((((((.((..(((....(((((((.....)))))))......((((((((......))))))))...-........)))..)))))))).. ( -35.60, z-score = -3.97, R) >droEre2.scaffold_4690 15034838 83 + 18748788 ---------GGUAAAAUGGCUGGGAAACUUUCCCGGCAAACACUGGGAAAUGAGGCAAUUUCCCAAAAACUCAGAACUUCGAGCCAAGCAAU ---------(((......(((((((.....)))))))......((((((((......)))))))).................)))....... ( -26.10, z-score = -2.66, R) >droYak2.chrX 2066823 80 + 21770863 ------------AUAAUGGCUGGGAAACUUUCCCGGCAAACACUGGGAAAUGAGGCAAUUUCCCAAAAACUCAGAACUUGAAGCCAGGCAAU ------------......(((((((.....)))))))......((((((((......))))))))...........((((....)))).... ( -24.30, z-score = -2.29, R) >droSim1.chrX 4781709 91 + 17042790 UGCUUGUGCGGGAAAAUGGCUGGGAAACUUUCCCGGCAAACACUGGGAAAUGAGGCAAUUUCCCAAAA-CUCAGAACUUCGAGCCAAGCAAU ((((((.((..(((....(((((((.....)))))))......((((((((......))))))))...-........)))..)))))))).. ( -35.60, z-score = -3.97, R) >consensus _________GGGAAAAUGGCUGGGAAACUUUCCCGGCAAACACUGGGAAAUGAGGCAAUUUCCCAAAA_CUCAGAACUUCGAGCCAAGCAAU ................(((((((((.....)))).........((((((((......))))))))................)))))...... (-23.27 = -23.27 + 0.00)

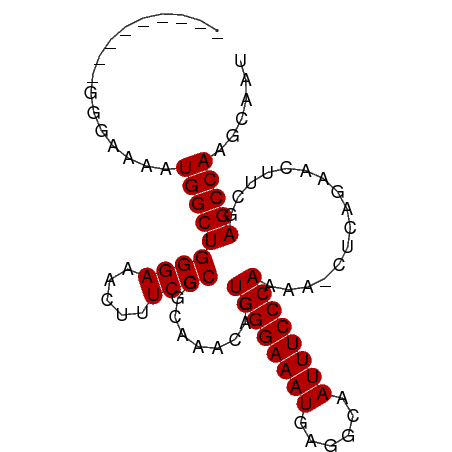
| Location | 6,056,614 – 6,056,705 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 88.38 |
| Shannon entropy | 0.17791 |
| G+C content | 0.46872 |
| Mean single sequence MFE | -26.91 |
| Consensus MFE | -21.77 |
| Energy contribution | -21.77 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.834905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 6056614 91 - 22422827 AUUGCUUGGCUCGAAGUUCUGAG-UUUUGGGAAAUUGCCUCAUUUCCCAGUGUUUGCCGGGAAAGUUUCCCAGCCAUUUUCCCGCACAAGCA ..(((((((((((......))))-).(((((((((......)))))))))....(((.((((((((.........))))))))))))))))) ( -31.00, z-score = -2.66, R) >droEre2.scaffold_4690 15034838 83 - 18748788 AUUGCUUGGCUCGAAGUUCUGAGUUUUUGGGAAAUUGCCUCAUUUCCCAGUGUUUGCCGGGAAAGUUUCCCAGCCAUUUUACC--------- ......(((((((......))).....((((((((.......((((((.((....)).)))))))))))))))))).......--------- ( -22.41, z-score = -1.11, R) >droYak2.chrX 2066823 80 - 21770863 AUUGCCUGGCUUCAAGUUCUGAGUUUUUGGGAAAUUGCCUCAUUUCCCAGUGUUUGCCGGGAAAGUUUCCCAGCCAUUAU------------ ......((((((((.....))))....((((((((.......((((((.((....)).))))))))))))))))))....------------ ( -23.21, z-score = -1.58, R) >droSim1.chrX 4781709 91 - 17042790 AUUGCUUGGCUCGAAGUUCUGAG-UUUUGGGAAAUUGCCUCAUUUCCCAGUGUUUGCCGGGAAAGUUUCCCAGCCAUUUUCCCGCACAAGCA ..(((((((((((......))))-).(((((((((......)))))))))....(((.((((((((.........))))))))))))))))) ( -31.00, z-score = -2.66, R) >consensus AUUGCUUGGCUCGAAGUUCUGAG_UUUUGGGAAAUUGCCUCAUUUCCCAGUGUUUGCCGGGAAAGUUUCCCAGCCAUUUUCCC_________ ......(((((...............(((((((((......)))))))))........((((.....)))))))))................ (-21.77 = -21.77 + 0.00)

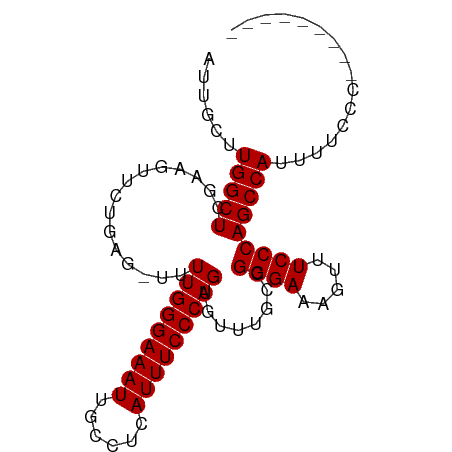
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:19:50 2011