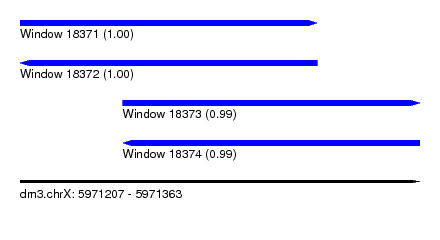
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,971,207 – 5,971,363 |
| Length | 156 |
| Max. P | 0.997642 |

| Location | 5,971,207 – 5,971,323 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 77.03 |
| Shannon entropy | 0.37910 |
| G+C content | 0.42263 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -24.50 |
| Energy contribution | -25.02 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.11 |
| SVM RNA-class probability | 0.997468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5971207 116 + 22422827 UAAAAACAAAAAUGUAAUAAGAAAAUCCAAAGAUAUGUCCGUGAUUUCU-CUAUGUGCAUCAGACUUUGGCGCUGAUUUACCGGAUCGCCAAAGUCCGUCGCCGUGAAAGUCAAUUG .........................................((((((.(-(.(((.((..(.(((((((((((((......)))..)))))))))).)..))))))))))))).... ( -27.50, z-score = -2.60, R) >droEre2.scaffold_4690 14950264 93 + 18748788 ------------------------UUAUUAUGACAAAGCCUUAAUUUUUUCUAUGUGCAUCAGACUUUGGCGCUGAUUUAUCGGAUCGCCAAAGUUCGUUGCCGUGAAAGUCAAUUG ------------------------......(((((((.......)))((((.(((.(((.(..(((((((((((((....))))..)))))))))..).)))))))))))))).... ( -26.80, z-score = -3.16, R) >droYak2.chrX 1980696 93 + 21770863 -----------------------AUACCAAUGAUAUGGCCCUGAUUUCG-CAAUGUGCAUCAGACUUUGGCGCUGAUUUAGCGGGUCGCCAAAGUCCGUUGCCGUGAAAGUCAAUCG -----------------------........(((.((((.....(((((-(...(.(((.(.(((((((((((((......)))..)))))))))).).))))))))))))))))). ( -28.70, z-score = -1.53, R) >droSec1.super_4 5383884 109 - 6179234 -------UAAAAAGUAAUGAGAAAAUCCAAAGAUAUAUCCGUGAUUUCU-CUACGUGCGUCAGACUUUGGCGCUGAUUUACCGAAUCGCCAAAGUCCGUCGCCGUGAAAGUCAAUUG -------...........((((((...((..((....))..)).)))))-)((((.(((.(.((((((((((.((......))...)))))))))).).)))))))........... ( -30.40, z-score = -3.69, R) >droSim1.chrX 4695510 109 + 17042790 -------UAAAGAGUAAUGAGAAAAUCCAUAGAUAUAUCCGUGAUUUUU-CUAUGUGCAUCAGACUUUGGCGCUGAUUUACCGGAUCGCCAAAGUCCGUCGCCGUGAAAGUCAAUUG -------..........((((((((..((..((....))..))..))))-)((((.((..(.(((((((((((((......)))..)))))))))).)..))))))....))).... ( -29.70, z-score = -2.79, R) >consensus ________AAA__GUAAU_AGAAAAUCCAAAGAUAUAUCCGUGAUUUCU_CUAUGUGCAUCAGACUUUGGCGCUGAUUUACCGGAUCGCCAAAGUCCGUCGCCGUGAAAGUCAAUUG .........................................((((((....((((.(((.(.(((((((((((((......)))..)))))))))).).))))))).)))))).... (-24.50 = -25.02 + 0.52)


| Location | 5,971,207 – 5,971,323 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.03 |
| Shannon entropy | 0.37910 |
| G+C content | 0.42263 |
| Mean single sequence MFE | -29.11 |
| Consensus MFE | -22.88 |
| Energy contribution | -22.68 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.15 |
| SVM RNA-class probability | 0.997642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5971207 116 - 22422827 CAAUUGACUUUCACGGCGACGGACUUUGGCGAUCCGGUAAAUCAGCGCCAAAGUCUGAUGCACAUAG-AGAAAUCACGGACAUAUCUUUGGAUUUUCUUAUUACAUUUUUGUUUUUA ...............(((.((((((((((((..............)))))))))))).)))....((-((((.(((.(((....))).))).))))))................... ( -29.34, z-score = -2.50, R) >droEre2.scaffold_4690 14950264 93 - 18748788 CAAUUGACUUUCACGGCAACGAACUUUGGCGAUCCGAUAAAUCAGCGCCAAAGUCUGAUGCACAUAGAAAAAAUUAAGGCUUUGUCAUAAUAA------------------------ ........((((...(((.((.(((((((((....((....))..))))))))).)).))).....))))..((((.(((...))).))))..------------------------ ( -20.00, z-score = -1.47, R) >droYak2.chrX 1980696 93 - 21770863 CGAUUGACUUUCACGGCAACGGACUUUGGCGACCCGCUAAAUCAGCGCCAAAGUCUGAUGCACAUUG-CGAAAUCAGGGCCAUAUCAUUGGUAU----------------------- ...((((.((((.(((((.((((((((((((..............)))))))))))).)))....))-.)))))))).((((......))))..----------------------- ( -31.14, z-score = -2.81, R) >droSec1.super_4 5383884 109 + 6179234 CAAUUGACUUUCACGGCGACGGACUUUGGCGAUUCGGUAAAUCAGCGCCAAAGUCUGACGCACGUAG-AGAAAUCACGGAUAUAUCUUUGGAUUUUCUCAUUACUUUUUA------- ............((((((.((((((((((((..............)))))))))))).))).))).(-(((((((.((((......)))))).))))))...........------- ( -35.14, z-score = -4.48, R) >droSim1.chrX 4695510 109 - 17042790 CAAUUGACUUUCACGGCGACGGACUUUGGCGAUCCGGUAAAUCAGCGCCAAAGUCUGAUGCACAUAG-AAAAAUCACGGAUAUAUCUAUGGAUUUUCUCAUUACUCUUUA------- ...............(((.((((((((((((..............)))))))))))).)))....((-((((.(((..((....))..))).))))))............------- ( -29.94, z-score = -3.07, R) >consensus CAAUUGACUUUCACGGCGACGGACUUUGGCGAUCCGGUAAAUCAGCGCCAAAGUCUGAUGCACAUAG_AGAAAUCACGGAUAUAUCUUUGGAUUUUCU_AUUAC__UUU________ ...............(((.((((((((((((..............)))))))))))).)))........................................................ (-22.88 = -22.68 + -0.20)

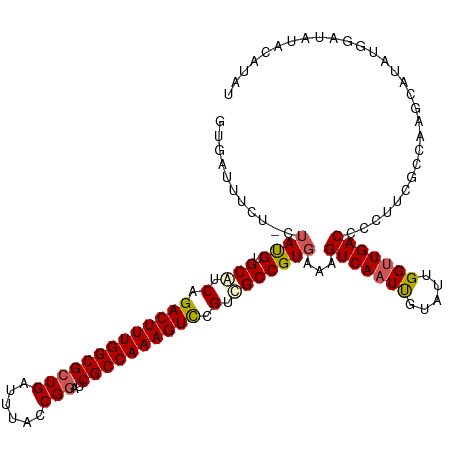
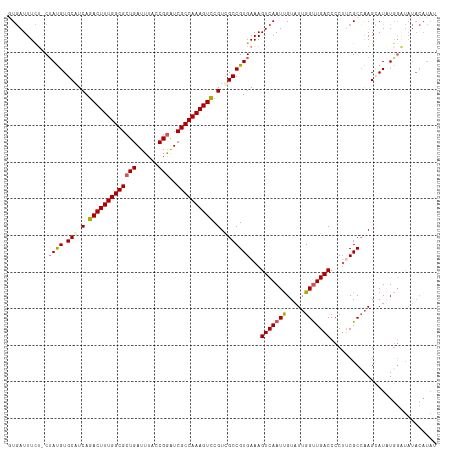
| Location | 5,971,247 – 5,971,363 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 86.43 |
| Shannon entropy | 0.24520 |
| G+C content | 0.45825 |
| Mean single sequence MFE | -36.52 |
| Consensus MFE | -27.98 |
| Energy contribution | -28.34 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.990707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5971247 116 + 22422827 GUGAUUUCU-CUAUGUGCAUCAGACUUUGGCGCUGAUUUACCGGAUCGCCAAAGUCCGUCGCCGUGAAAGUCAAUUGUAUUGGUUGACCCCUUCGCAAAGUAUCUGGAUAUACAUAU .........-.((((((.(((.(((((((((((((......)))..)))))))))).......(((((.(((((((.....)))))))...)))))..........))).)))))). ( -34.70, z-score = -2.53, R) >droEre2.scaffold_4690 14950280 107 + 18748788 UUAAUUUUUUCUAUGUGCAUCAGACUUUGGCGCUGAUUUAUCGGAUCGCCAAAGUUCGUUGCCGUGAAAGUCAAUUGUAUUGGUUGACCCCACCGCCAAGUAUAUAC---------- ...........(((((((....((((((((((((((....))))..))))))))))....((.(((...(((((((.....)))))))..))).))...))))))).---------- ( -32.40, z-score = -3.58, R) >droYak2.chrX 1980713 116 + 21770863 CUGAUUUCG-CAAUGUGCAUCAGACUUUGGCGCUGAUUUAGCGGGUCGCCAAAGUCCGUUGCCGUGAAAGUCAAUCGUAUUGGCUGACCCCUGCGCCAAGCAUCUGGUUAUACAUAU .........-..(((((.((((((.((((((((......((.((((((((((...(.((((.(......).)))).)..))))).)))))))))))))))..))))))..))))).. ( -38.40, z-score = -1.77, R) >droSec1.super_4 5383917 116 - 6179234 GUGAUUUCU-CUACGUGCGUCAGACUUUGGCGCUGAUUUACCGAAUCGCCAAAGUCCGUCGCCGUGAAAGUCAAUUGUAUGGGUUGACCCCUUCGCCGAGAAUAUGGAUAUACAUAU .....((((-(((((.(((.(.((((((((((.((......))...)))))))))).).)))))))...(((((((.....))))))).........)))))((((......)))). ( -37.50, z-score = -2.81, R) >droSim1.chrX 4695543 116 + 17042790 GUGAUUUUU-CUAUGUGCAUCAGACUUUGGCGCUGAUUUACCGGAUCGCCAAAGUCCGUCGCCGUGAAAGUCAAUUGUAUUGGUUGACCCCUUCGCCAAGCAUAUGGAUAUACAUAU (((.....(-((((((((....(((((((((((((......)))..)))))))))).......(((((.(((((((.....)))))))...)))))...)))))))))....))).. ( -39.60, z-score = -3.89, R) >consensus GUGAUUUCU_CUAUGUGCAUCAGACUUUGGCGCUGAUUUACCGGAUCGCCAAAGUCCGUCGCCGUGAAAGUCAAUUGUAUUGGUUGACCCCUUCGCCAAGCAUAUGGAUAUACAUAU ...........((((.(((.(.(((((((((((((......)))..)))))))))).).)))))))...(((((((.....)))))))............................. (-27.98 = -28.34 + 0.36)

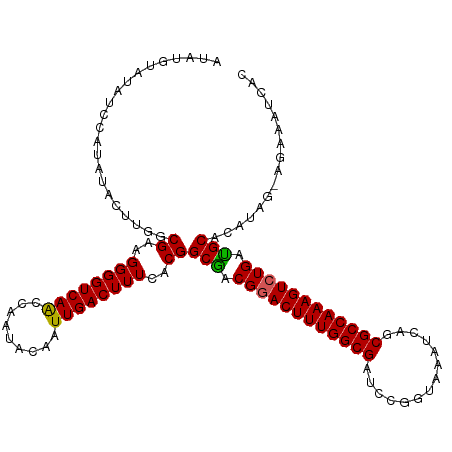
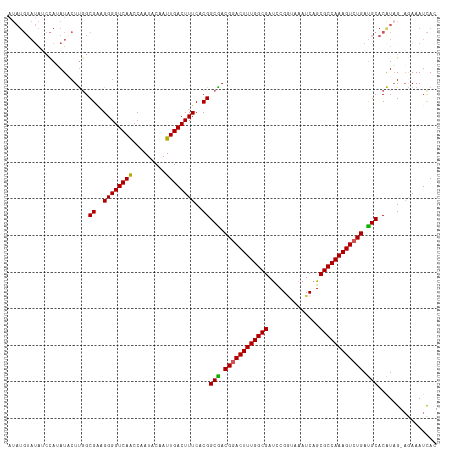
| Location | 5,971,247 – 5,971,363 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 86.43 |
| Shannon entropy | 0.24520 |
| G+C content | 0.45825 |
| Mean single sequence MFE | -35.24 |
| Consensus MFE | -29.82 |
| Energy contribution | -29.46 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.70 |
| SVM RNA-class probability | 0.994479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5971247 116 - 22422827 AUAUGUAUAUCCAGAUACUUUGCGAAGGGGUCAACCAAUACAAUUGACUUUCACGGCGACGGACUUUGGCGAUCCGGUAAAUCAGCGCCAAAGUCUGAUGCACAUAG-AGAAAUCAC .(((((................((..((((((((.........))))))))..))(((.((((((((((((..............)))))))))))).))))))))(-(....)).. ( -34.64, z-score = -2.94, R) >droEre2.scaffold_4690 14950280 107 - 18748788 ----------GUAUAUACUUGGCGGUGGGGUCAACCAAUACAAUUGACUUUCACGGCAACGAACUUUGGCGAUCCGAUAAAUCAGCGCCAAAGUCUGAUGCACAUAGAAAAAAUUAA ----------((((.......((.((((((((((.........))))).))))).))..((.(((((((((....((....))..))))))))).))))))................ ( -28.50, z-score = -2.33, R) >droYak2.chrX 1980713 116 - 21770863 AUAUGUAUAACCAGAUGCUUGGCGCAGGGGUCAGCCAAUACGAUUGACUUUCACGGCAACGGACUUUGGCGACCCGCUAAAUCAGCGCCAAAGUCUGAUGCACAUUG-CGAAAUCAG ...(((((...(((((..((((((((((((((.(((((...((.......)).((....))....)))))))))).))......))))))).)))))))))).....-......... ( -39.70, z-score = -2.87, R) >droSec1.super_4 5383917 116 + 6179234 AUAUGUAUAUCCAUAUUCUCGGCGAAGGGGUCAACCCAUACAAUUGACUUUCACGGCGACGGACUUUGGCGAUUCGGUAAAUCAGCGCCAAAGUCUGACGCACGUAG-AGAAAUCAC .((((......))))(((((.(((..(((.....)))..................(((.((((((((((((..............)))))))))))).))).))).)-))))..... ( -39.34, z-score = -3.75, R) >droSim1.chrX 4695543 116 - 17042790 AUAUGUAUAUCCAUAUGCUUGGCGAAGGGGUCAACCAAUACAAUUGACUUUCACGGCGACGGACUUUGGCGAUCCGGUAAAUCAGCGCCAAAGUCUGAUGCACAUAG-AAAAAUCAC .(((((....(((......)))((..((((((((.........))))))))..))(((.((((((((((((..............)))))))))))).)))))))).-......... ( -34.04, z-score = -2.47, R) >consensus AUAUGUAUAUCCAUAUACUUGGCGAAGGGGUCAACCAAUACAAUUGACUUUCACGGCGACGGACUUUGGCGAUCCGGUAAAUCAGCGCCAAAGUCUGAUGCACAUAG_AGAAAUCAC ......................((..((((((((.........))))))))..))(((.((((((((((((..............)))))))))))).)))................ (-29.82 = -29.46 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:19:36 2011