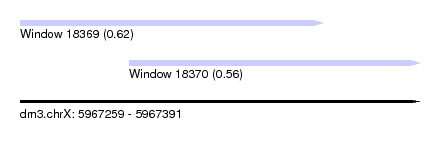
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,967,259 – 5,967,391 |
| Length | 132 |
| Max. P | 0.622837 |

| Location | 5,967,259 – 5,967,359 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 98.67 |
| Shannon entropy | 0.01837 |
| G+C content | 0.53333 |
| Mean single sequence MFE | -27.07 |
| Consensus MFE | -27.29 |
| Energy contribution | -27.07 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -0.86 |
| Structure conservation index | 1.01 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.622837 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
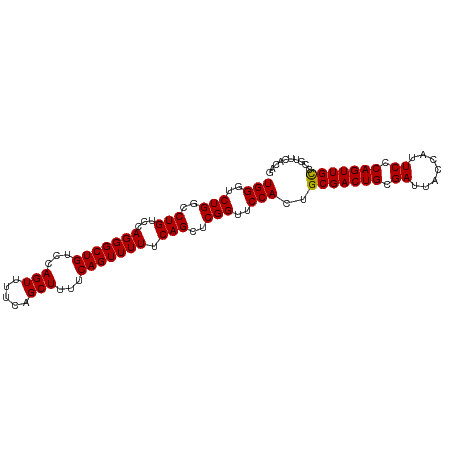
>dm3.chrX 5967259 100 + 22422827 UGGGUCUGGCCUGUCCAGGGCUGUCCAGUUUUCAGCUUUUCAGUUUUUCAGCUCGGUUCCACUGCGACUGCGAUUACCAUUCCCAGUUGCCCGUUCAUAG (((..(((..(((...(((((((...(((.....)))...))))))).)))..)))..)))..(((((((.((.......)).))))))).......... ( -27.70, z-score = -1.12, R) >droSec1.super_4 5379858 100 - 6179234 UGGGUCUGGCCUGUCCAGGGCUGUCCAGUUUUCAGCUUUUCAGUUUUUCAGCUCGGUUCCACUGCGACUGCGAUUACCAUUCCCAGUUGCCCGUUCACAG (((..(((..(((...(((((((...(((.....)))...))))))).)))..)))..)))..(((((((.((.......)).))))))).......... ( -27.70, z-score = -0.91, R) >droSim1.chrX 4691392 100 + 17042790 UGGGUCUGGCCUGUCCAGGGCUGUCCAGUUUUCAGCUUUUCAGUUUUUCAGCUCGGUUCCACUGCGACUGCGAUUACCAUUCCCAGUUGUCCGUUCACAG (((..(((..(((...(((((((...(((.....)))...))))))).)))..)))..)))..(((((((.((.......)).))))))).......... ( -25.80, z-score = -0.55, R) >consensus UGGGUCUGGCCUGUCCAGGGCUGUCCAGUUUUCAGCUUUUCAGUUUUUCAGCUCGGUUCCACUGCGACUGCGAUUACCAUUCCCAGUUGCCCGUUCACAG (((..(((..(((...(((((((...(((.....)))...))))))).)))..)))..)))..(((((((.((.......)).))))))).......... (-27.29 = -27.07 + -0.22)

| Location | 5,967,295 – 5,967,391 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 93.59 |
| Shannon entropy | 0.08609 |
| G+C content | 0.49306 |
| Mean single sequence MFE | -12.57 |
| Consensus MFE | -12.45 |
| Energy contribution | -12.23 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.79 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.559675 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
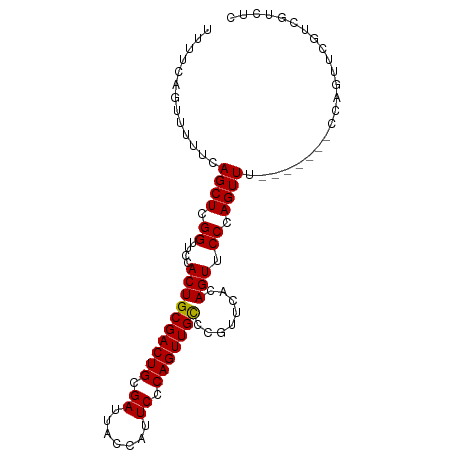
>dm3.chrX 5967295 96 + 22422827 UUUUCAGUUUUUCAGCUCGGUUCCACUGCGACUGCGAUUACCAUUCCCAGUUGCCCGUUCAUAGUUCCCAGUUUUCAGUUUCCAGUUCGUCGUCUC .............((((.((..(.((.(((((((.((.......)).)))))))..)).....)..)).))))....................... ( -12.70, z-score = -0.91, R) >droSec1.super_4 5379894 89 - 6179234 UUUUCAGUUUUUCAGCUCGGUUCCACUGCGACUGCGAUUACCAUUCCCAGUUGCCCGUUCACAGUUCCCAGUUU-------CCAGUUCGUCGUCUC .............((((.((..(....(((((((.((.......)).)))))))..(....).)..)).)))).-------............... ( -12.90, z-score = -0.83, R) >droSim1.chrX 4691428 89 + 17042790 UUUUCAGUUUUUCAGCUCGGUUCCACUGCGACUGCGAUUACCAUUCCCAGUUGUCCGUUCACAGUUCCCAGUUU-------CCAGUUCGUCGUCUC .............((((.((....((((.((.((.(((.((........)).))))).)).)))).)).)))).-------............... ( -12.10, z-score = -0.63, R) >consensus UUUUCAGUUUUUCAGCUCGGUUCCACUGCGACUGCGAUUACCAUUCCCAGUUGCCCGUUCACAGUUCCCAGUUU_______CCAGUUCGUCGUCUC .............((((.((....((((((((((.((.......)).)))))))........))).)).))))....................... (-12.45 = -12.23 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:19:33 2011