| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,959,125 – 5,959,193 |
| Length | 68 |
| Max. P | 0.918051 |

| Location | 5,959,125 – 5,959,193 |
|---|---|
| Length | 68 |
| Sequences | 15 |
| Columns | 77 |
| Reading direction | forward |
| Mean pairwise identity | 72.27 |
| Shannon entropy | 0.64118 |
| G+C content | 0.43829 |
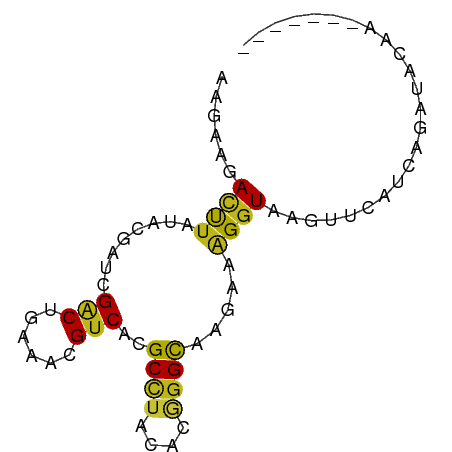
| Mean single sequence MFE | -13.91 |
| Consensus MFE | -7.62 |
| Energy contribution | -7.13 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.00 |
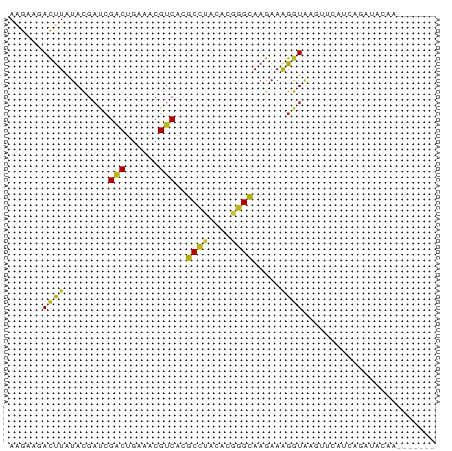
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.918051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
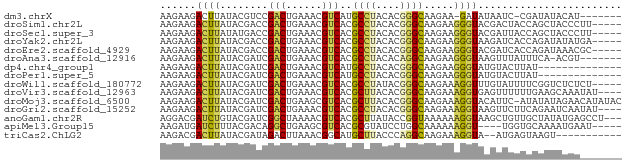
>dm3.chrX 5959125 68 + 22422827 AAGAAGACUUAUACGUCCGACUGAAACGUCAUGCCUACACGGGCAAGAA-GAUAUAAUC-CGAUAUACAU------- .....(((......))).(((......))).(((((....)))))....-.........-..........------- ( -10.40, z-score = -0.58, R) >droSim1.chr2L 15266097 72 - 22036055 AAGAAGACUUAUACGACCGACUGAAACGUCACGCCUACACGGGCAAGAAGGGUACGACUACCAGCUACCCUU----- .......(((........(((......)))..((((....)))))))((((((((........).)))))))----- ( -16.60, z-score = -1.71, R) >droSec1.super_3 4713837 72 - 7220098 AAGAAGACUUAUAUGACCGACUGAAACGUCACGCCUACACGGGCAAGAAGGGUACGAUUACCAGCUACCCUU----- .......(((........(((......)))..((((....)))))))((((((((........).)))))))----- ( -16.60, z-score = -1.89, R) >droYak2.chr2L 5935888 72 + 22324452 AAGAAGACUUAUACGACCGACUGAAACGUCACGCCUACACGGGCAAGAAGGGUAAGAUCACCAGAUAUAUGA----- .....(((((((....(((((......)))..((((....)))).....))))))).)).............----- ( -13.00, z-score = -0.88, R) >droEre2.scaffold_4929 10751753 72 - 26641161 AAGAAGACUUAUACGACCGACUGAAACGUCACGCCUACACGGGCAAGAAGGGUACGAUCACCAGAUAAACGC----- .............((((((((......)))..((((....))))......))).))................----- ( -13.30, z-score = -0.69, R) >droAna3.scaffold_12916 16127294 69 - 16180835 AAGAAGACUUAUACGAUCGACUGAAACGUCACGCCUACACAGGCAAGAAGGGUAAGUUUAUUUCA-ACGU------- ....(((((((..(..(((((......)))..((((....))))..)).)..)))))))......-....------- ( -15.40, z-score = -1.82, R) >dp4.chr4_group1 3765827 63 + 5278887 AAGAAGACUUAUACGAUCGACUGAAACGUCAUGCCUACACGGGCAAGAAGGGUAUGUACUUAU-------------- ...(((...(((((....(((......))).(((((....)))))......)))))..)))..-------------- ( -12.80, z-score = -1.19, R) >droPer1.super_5 5303473 63 - 6813705 AAGAAGACUUAUACGAUCGACUGAAACGUCAUGCCUACACGGGCAAGAAGGGUAUGUACUUAU-------------- ...(((...(((((....(((......))).(((((....)))))......)))))..)))..-------------- ( -12.80, z-score = -1.19, R) >droWil1.scaffold_180772 7270548 72 + 8906247 AAGAAGACUUAUACGAUCGACUGAAACGUCACGCCUAUACGGGCAAGAAAGGUUUGUAUUUUCGGUCUCUCU----- .((((((((.(((((((((((......)))..((((....))))......)).))))))....))))).)))----- ( -18.00, z-score = -1.30, R) >droVir3.scaffold_12963 16792177 73 - 20206255 AAGAAGACUUAUACGAUCGACUGAAACGUCACGCUUACACGGGCAAGAAAGGUGAGUUUUUUGAAGCAAAUAU---- .(((((((((((....(((((......)))..(((......)))..))...)))))))))))...........---- ( -15.20, z-score = -1.51, R) >droMoj3.scaffold_6500 18564640 76 - 32352404 AAGAAGACUUAUACGAUCGACUGAAGCGUCACGCUUACACGGGCAAGAAAGGUACAUUC-AUAUAUAGAACAUAUAC ..(((.((((........(((......)))..(((......))).....))))...)))-................. ( -9.40, z-score = -0.12, R) >droGri2.scaffold_15252 14215504 73 - 17193109 AAGAAGACUUAUACGAUCGACUGAAACGUCACGCCUACACGGGCAAGAAAGGUAAGUUCUUCAGAAUCAAUAU---- ..((((((((((....(((((......)))..((((....))))..))...))))).)))))...........---- ( -18.60, z-score = -2.83, R) >anoGam1.chr2R 26440537 74 + 62725911 AGGACGAUCUGUACGAUCGGCUAAAACGUCACGCUUAUACCGGUAAAAAAGGUAAGCUGUUGCUAUAUGAGCCU--- .....((((.....))))((((.....((.(((((..((((.........))))))).)).))......)))).--- ( -17.20, z-score = -0.55, R) >apiMel3.Group15 3610558 68 - 7856270 AAGAUGAUCUUUACGACAGGCUGAAGCGUCACGCGUAUCCUGGCAAAAAAGGU----UGGUGCAAAAUGAAU----- .....(((((((....((((.....(((...)))....)))).....))))))----)..............----- ( -11.80, z-score = 0.75, R) >triCas2.ChLG2 2058366 64 + 12900155 AAGACGACUUAUACGAUAGACUUAAACGGCAUGCUUACCCAGGCAAGAAAGGUA--AUGAGUAAGU----------- ......((((((.((....((((......(.(((((....))))).)..)))).--.)).))))))----------- ( -7.60, z-score = 0.48, R) >consensus AAGAAGACUUAUACGAUCGACUGAAACGUCACGCCUACACGGGCAAGAAAGGUAAGUUCAUCAGAUACAA_______ ......((((........(((......)))..((((....)))).....))))........................ ( -7.62 = -7.13 + -0.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:19:32 2011