| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,952,113 – 5,952,187 |
| Length | 74 |
| Max. P | 0.963204 |

| Location | 5,952,113 – 5,952,187 |
|---|---|
| Length | 74 |
| Sequences | 4 |
| Columns | 81 |
| Reading direction | forward |
| Mean pairwise identity | 70.57 |
| Shannon entropy | 0.43464 |
| G+C content | 0.37288 |
| Mean single sequence MFE | -15.50 |
| Consensus MFE | -5.85 |
| Energy contribution | -7.10 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.953193 |
| Prediction | RNA |
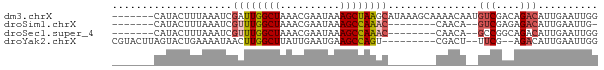
Download alignment: ClustalW | MAF
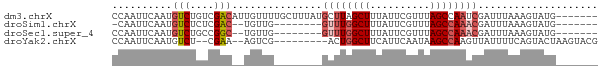
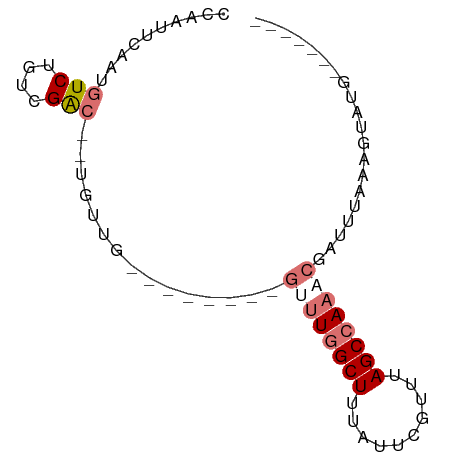
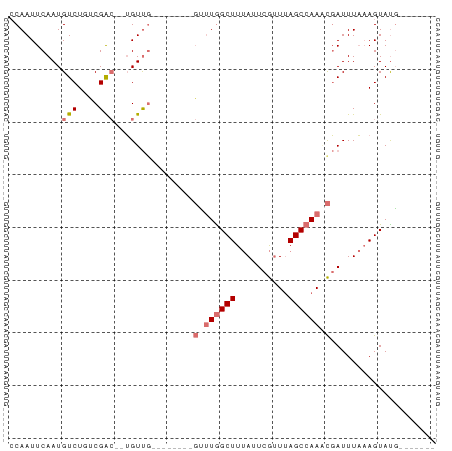
>dm3.chrX 5952113 74 + 22422827 CCAAUUCAAUGUCUGUCGACAUUGUUUUGCUUUAUGCUUAGCUUUAUUCGUUUAGCCAAUCGAUUUAAAGUAUG------- .(((..(((((((....)))))))..)))...........((((((.(((..........)))..))))))...------- ( -15.60, z-score = -3.02, R) >droSim1.chrX 4677218 63 + 17042790 -CAAUUCAAUGUCUCUCGAC--UGUUG--------GUUUGGCUUUAUUCGUUUAGCCAAACGAUUUAAAGUAUG------- -(((..(((((((....)))--.))))--------..)))((((((.((((((....))))))..))))))...------- ( -15.70, z-score = -3.30, R) >droSec1.super_4 5366191 64 - 6179234 CCAAUUCAAUGUCUGCCGGC--UGUUG--------GUUUGGCUUUAUUCGUUUAGCCAAACGAUUUAAAGUAUG------- .(((..(((((((....)))--.))))--------..)))((((((.((((((....))))))..))))))...------- ( -15.40, z-score = -1.87, R) >droYak2.chrX 14810895 68 + 21770863 CCAAUUCAAUGUCU--CGAA--AGUCG---------ACUGGCUUCAUUCAAUAAGCCAAGUUAUUUUCAGUACUAAGUACG ..............--.(((--(((.(---------((((((((........))))).)))))))))).((((...)))). ( -15.30, z-score = -3.68, R) >consensus CCAAUUCAAUGUCUGUCGAC__UGUUG________GUUUGGCUUUAUUCGUUUAGCCAAACGAUUUAAAGUAUG_______ ...................................((((((((..........)))))))).................... ( -5.85 = -7.10 + 1.25)
| Location | 5,952,113 – 5,952,187 |
|---|---|
| Length | 74 |
| Sequences | 4 |
| Columns | 81 |
| Reading direction | reverse |
| Mean pairwise identity | 70.57 |
| Shannon entropy | 0.43464 |
| G+C content | 0.37288 |
| Mean single sequence MFE | -13.05 |
| Consensus MFE | -7.45 |
| Energy contribution | -8.32 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.963204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
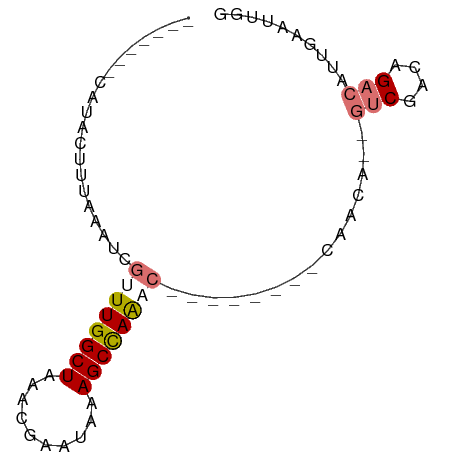
>dm3.chrX 5952113 74 - 22422827 -------CAUACUUUAAAUCGAUUGGCUAAACGAAUAAAGCUAAGCAUAAAGCAAAACAAUGUCGACAGACAUUGAAUUGG -------....(((((......((((((..........))))))...)))))(((..(((((((....)))))))..))). ( -15.20, z-score = -3.30, R) >droSim1.chrX 4677218 63 - 17042790 -------CAUACUUUAAAUCGUUUGGCUAAACGAAUAAAGCCAAAC--------CAACA--GUCGAGAGACAUUGAAUUG- -------.....(((((...((((((((..........))))))))--------.....--(((....))).)))))...- ( -15.40, z-score = -4.15, R) >droSec1.super_4 5366191 64 + 6179234 -------CAUACUUUAAAUCGUUUGGCUAAACGAAUAAAGCCAAAC--------CAACA--GCCGGCAGACAUUGAAUUGG -------.............((((((((..........))))))))--------.....--.................... ( -10.00, z-score = -0.68, R) >droYak2.chrX 14810895 68 - 21770863 CGUACUUAGUACUGAAAAUAACUUGGCUUAUUGAAUGAAGCCAGU---------CGACU--UUCG--AGACAUUGAAUUGG ........((..(((((.....(((((((........))))))).---------....)--))))--..)).......... ( -11.60, z-score = -0.65, R) >consensus _______CAUACUUUAAAUCGUUUGGCUAAACGAAUAAAGCCAAAC________CAACA__GUCGACAGACAUUGAAUUGG ....................((((((((..........))))))))...............(((....))).......... ( -7.45 = -8.32 + 0.88)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:19:31 2011