| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,939,960 – 5,940,065 |
| Length | 105 |
| Max. P | 0.785000 |

| Location | 5,939,960 – 5,940,065 |
|---|---|
| Length | 105 |
| Sequences | 14 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 73.75 |
| Shannon entropy | 0.56297 |
| G+C content | 0.54614 |
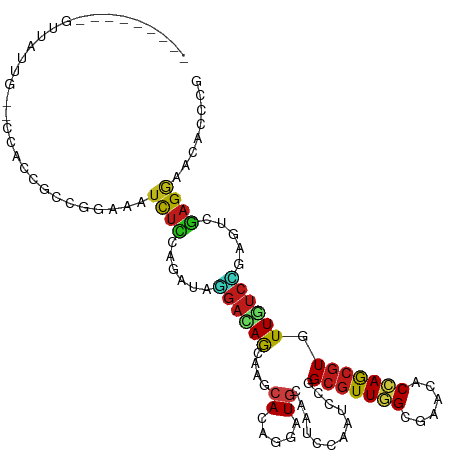
| Mean single sequence MFE | -32.64 |
| Consensus MFE | -12.44 |
| Energy contribution | -12.35 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.63 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.785000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
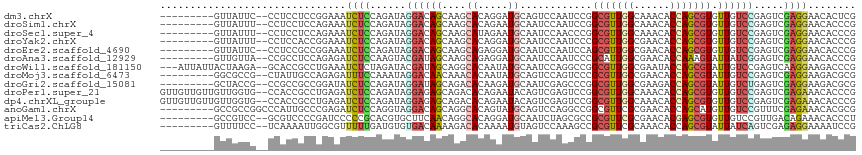
>dm3.chrX 5939960 105 - 22422827 ---------GUUAUUC--CCUCCUCCGGAAAUCUCCAGAUAGGACAGCAAGCACAGGAUGCAGUCCAAUCCGGCGUUGGCAAACACCAGCGUGUUGUCCGAGUCGAGGAACACUCG ---------.......--..(((((.(((....))).(((.((((((((.((...((((........)))).))(((((......))))).))))))))..))))))))....... ( -38.30, z-score = -3.40, R) >droSim1.chrX 4667514 105 - 17042790 ---------GUUAUUU--CCUCCUCCAGAAAUCUCCAGAUAGGACAGCAAGCACAGAAUGCAAUCCAAUCCGGCGUUGGCAAACACCAGCGUGUUGUCCGAGUCGAGGAACACCCG ---------.....((--(((((((.....(((....))).(((((((..(((.....)))...........(((((((......)))))))))))))))))..))))))...... ( -33.30, z-score = -3.60, R) >droSec1.super_4 5354105 105 + 6179234 ---------GUUAUUU--CCUCCUCCAGAAAUCUCCAGAUAGGACAGCAAGCAUAGAAUGCAAUCCAACCCGGCGUUGGCAAACACCAGCGUGUUGUCCGAGUCGAGGAACACCCG ---------.....((--(((((((.....(((....))).(((((((..((((...))))...........(((((((......)))))))))))))))))..))))))...... ( -33.70, z-score = -3.93, R) >droYak2.chrX 14798357 105 - 21770863 ---------GUUAUUU--CCUCCACCGGAAAUCUCCAGAUAGGACAGCAAGCACAGGAUGCAAUCCAAUCCCGCGUUGGCGAACACCAGCGUGUUGUCCGAGUCGAGGAACACCCG ---------.....((--((((....(((....))).(((.(((((((.......((((........)))).(((((((......))))))))))))))..)))))))))...... ( -37.70, z-score = -3.92, R) >droEre2.scaffold_4690 3274962 105 - 18748788 ---------GUUAUUC--CCUCCGCCGGAAAUCUCCAGAUAGGACAGCAAGCAGAGGAUGCAAUCCAAUCCAGCGUUGGCGAACACCAGCGUGUUGUCCGAGUCGAGGAACACCCG ---------.......--((((....(((....))).(((.(((((((.......((((........)))).(((((((......))))))))))))))..)))))))........ ( -36.50, z-score = -3.18, R) >droAna3.scaffold_12929 247586 105 - 3277472 ---------GUUGUUA--CCGCCUCCAGAGAUCUCCAAGUACGAUAGCAAGCAGAGGAUGCAAUCCAAUCCCGCAUUGGCGAACACCAAAGUAUUAUCGGAGUCGAGGAACACCCG ---------.......--...((((....((.((((.(((((........((.(.((((........))))))).((((......)))).)))))...))))))))))........ ( -22.10, z-score = -0.26, R) >droWil1.scaffold_181150 3086585 111 + 4952429 ---AUUAUUACUAAGA--GCACCGCCUGAAAUCUCUAGAUACGAUAGCAGGCACAAUAUGCAAUCCAGGCCGGCGUUGGCGAAUACCAGCGUAUUGUCCGAGUCAAGGAAGACCCG ---.............--.....(((((..(((.........)))..)))))(((((((((.......((((....))))(.....).))))))))).((.(((......))).)) ( -26.90, z-score = -0.38, R) >droMoj3.scaffold_6473 100092 105 - 16943266 ---------GGCGCCG--CUAUUGCCAGAGAUUUCCAAAUAGGACAACAAACACAAUAUGCAGUCCAGUCCCGCGUUGGCGAACACCAGCGUAUUGUCCGAGUCGAGGAAGACGCG ---------((.((.(--((((((.........(((.....))).........))))).)).)))).(((..(((((((......)))))))....(((.......))).)))... ( -24.67, z-score = 0.35, R) >droGri2.scaffold_15081 2746820 105 - 4274704 ---------GCUACCG--CCGCCGCCGGAUAUCUCCAGAUAGGAUAGCAGACACAAGAUGCAAUCGAGCCCGGCGUUGGCGAAGACCAGCGUAUUGUCUGAGUCGAGGAAGACGCG ---------((...((--(((.((((((.((((....)))).(((.(((.........))).)))....)))))).)))))..(((((((.....).))).))).........)). ( -35.50, z-score = -1.28, R) >droPer1.super_21 1289528 114 + 1645244 GUUGUUGUUGUUGGUG--CCACCGCCUGAGAUCUCCAGAUAGGAGAGCAGACACAGAAUACAGUCGAGUCCGGCGUUGGCAAACACCAGCGUGUUGUCCGAGUCGAGAAACACCCG ...((((.((((..((--(((.((((.(((.(((((.....))))).).(((..........)))...)).)))).))))))))).))))(((((.((((...)).)))))))... ( -36.60, z-score = -1.45, R) >dp4.chrXL_group1e 7083537 114 + 12523060 GUUGUUGUUGUUGGUG--CCACCGCCUGAGAUCUCCAGAUAGGAGAGCAGACACAGAAUACAGUCGAGUCCGGCGUUGGCAAACACCAGCGUGUUGUCCGAGUCGAGAAACACCCG ...((((.((((..((--(((.((((.(((.(((((.....))))).).(((..........)))...)).)))).))))))))).))))(((((.((((...)).)))))))... ( -36.60, z-score = -1.45, R) >anoGam1.chrX 11395098 107 - 22145176 ---------GCCGCCGGCCCAUUGCCCGAGAUCUCCAGGUAGGACAGCAGGCACAGUAUGCAGUCCAGGCCGGCGUUCGCGAACACCAGCAUGUUGUCCGUUUCGAGAAACACGCG ---------((((((((((...((((.((....))..))))((((.(((.((...)).))).)))).))))))))(((.((((.((..((.....))..)))))).)))....)). ( -38.60, z-score = -1.64, R) >apiMel3.Group14 1943787 105 - 8318479 ---------GCCGUCC--GCGUCCCCGAUCCCCCGCACGUGCUUCAACAGGCACAGGAUGCAAUCUAGCGCCGCGUUCGCGAACACGAGCGUGUUGUCCGUUGACAGAAACACCCU ---------...(((.--((((((.((......))...(((((......))))).))))))......(((.((((((((......)))))))).))).....)))........... ( -31.90, z-score = -0.67, R) >triCas2.ChLG8 3598922 105 - 15773733 ---------GUUUUCC--UCAAAAUUGGCGUUUUUGAUGUGUGACAAAAGACACAAAAUGUAGUCCAAAGCCGCGUUCGCAAACACCAGCGUAUUAUCAGUCGAGAGGAAAAUCCG ---------(((((((--((....((((.((((.(((((((.(......(((((.....)).))).....))))).))).)))).)))).......((....)))))))))))... ( -24.60, z-score = -1.46, R) >consensus _________GUUAUUG__CCACCGCCGGAAAUCUCCAGAUAGGACAGCAAGCACAGGAUGCAAUCCAAUCCGGCGUUGGCGAACACCAGCGUGUUGUCCGAGUCGAGGAACACCCG ...............................((((......((((((....((.....))............(((((((......))))))).)))))).....))))........ (-12.44 = -12.35 + -0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:19:29 2011