
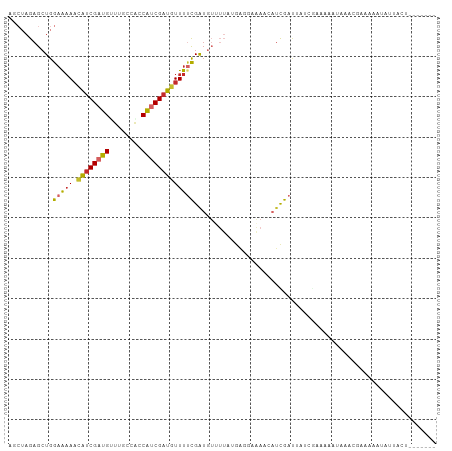
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,859,910 – 5,860,012 |
| Length | 102 |
| Max. P | 0.999829 |

| Location | 5,859,910 – 5,860,012 |
|---|---|
| Length | 102 |
| Sequences | 9 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 60.56 |
| Shannon entropy | 0.84136 |
| G+C content | 0.37068 |
| Mean single sequence MFE | -24.73 |
| Consensus MFE | -11.26 |
| Energy contribution | -11.37 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.50 |
| SVM RNA-class probability | 0.999829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
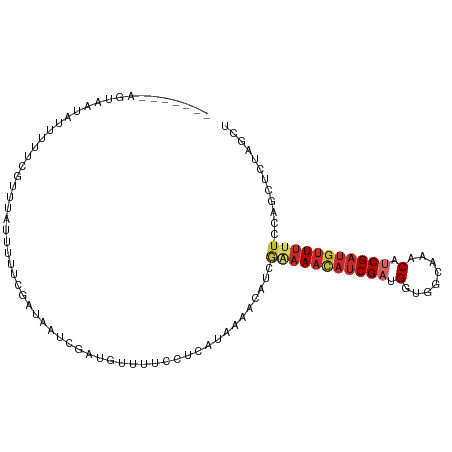
>dm3.chrX 5859910 102 + 22422827 AGCUAGAGCUGGAAAAACAUCGAUGCUCGUCGCCGUCGAUGUUUUCGAUGUUUUAUGGCGAGAACAUUGACUAUCGAAAAAUAAACGAAAAAUAUUACUCUU---- .....(((((.((......)).).)))).(((..(((((((((((((...........)))))))))))))...))).........................---- ( -24.40, z-score = -1.32, R) >droWil1.scaffold_181089 7826887 93 + 12369635 -GCUAGAGCUGGAAAAACAUCGAUGUUUGGGACCAUCGAUGUUUUCGAUGUUUUCAACGUCAAACAUCGAUGAAACAUCGAUGUUUCGA-UGUUU----------- -((....)).....(((((((((((((.(..(.((((((.....)))))).)..)))))))(((((((((((...))))))))))).))-)))))----------- ( -36.20, z-score = -4.73, R) >droSim1.chrX 4586294 102 + 17042790 AGCUAGAGCUGGAAAAACAGCGAUGCUUUCCACCGUCGAUGUUUUCGAUGUUUUAUGGCGAGAACAUUGACUAUCGAAAAAUAAACGAAAAAUAUUACUCUU---- .((....((((......))))...))((((....(((((((((((((...........)))))))))))))....)))).......................---- ( -25.80, z-score = -2.56, R) >droSec1.super_4 5274257 100 - 6179234 AGCUAGAGCUGGAAAAACAUCGAUGCUUACUGCCGUCGAUGUUUUCGAUGUUUUAUGGGGAGAACAUUGACUAUCGAAA--UAAACGAAAAAUAUUACUCUU---- ....((((.(((..((((((((((((.....).)))))))))))((((((((((......)))))))))))))(((...--....))).........)))).---- ( -25.40, z-score = -2.72, R) >droYak2.chrX 14713187 97 + 21770863 UGCAAGAGAUGGUAAAAUGUCGAUGUCUGCCCACAACGAUGUUUCCGAU-------GAUACUAUAAUCGAUUGUCGAAAAAA--AACAAAAUAAGAACUUACAAUU .......((..((.((((((((.(((......))).)))))))).((((-------.........))))))..)).......--...................... ( -13.40, z-score = -0.02, R) >droEre2.scaffold_4690 3197778 91 + 18748788 GGCUAGAGCUGGAAAAACAUCGAUGUUUGCCAGCAACGAUAUUUUUGG--------GCGACUAUAAUCGAUUAUCGAAAAAAGAAACGGAAUAACAAAC------- .......(((((..(((((....))))).)))))..(((((.((.(((--------....))).....)).))))).......................------- ( -16.70, z-score = -1.31, R) >dp4.Unknown_group_23 12088 85 + 67933 AUCUAGAGCUGGAGAAACAUCGAUGGUUGCGAACAUCGAUGUUUUCGAUGUUUUGAGAGCAAA----UGAAAUUAACCCAUUAACGGCC----------------- .......(((((((((.((((((((........)))))))))))))((((..((((...(...----.)...))))..))))..)))).----------------- ( -24.10, z-score = -2.67, R) >droPer1.super_0 2206017 99 + 11822988 AUAUAGAGCUGGAGAAACAUCGAUGUUUGCGGACAUCGAUGUUUACGAUGUUUUGGGAGCAAAACAUCGAUGUUUUUUAAAUAAAACUUAUAAACUUAU------- ...........((((((((((((((((....))))))))))))).((((((((((....))))))))))..(((((......))))).......)))..------- ( -32.70, z-score = -6.05, R) >droMoj3.scaffold_6500 31529585 99 + 32352404 ----AGAGCUGGAAUAUCAUCGAUGUUUACCACCAUCGAUGUUUUCGAUGUGUUUUGAAGAAAACAUCGAUGUUACAU-GUUGCGCGGAAUAUUUAAAACUAUU-- ----...((.(.((((((((((((((((.(.((((((((.....)))))).))......).)))))))))))....))-))).)))..................-- ( -23.90, z-score = -1.59, R) >consensus AGCUAGAGCUGGAAAAACAUCGAUGUUUGCCACCAUCGAUGUUUUCGAUGUUUUAUGAGGAAAACAUCGAUUAUCGAAAAAUAAACGAAAAAUAUUACU_______ ...........(((((.((((((((........)))))))))))))............................................................ (-11.26 = -11.37 + 0.10)


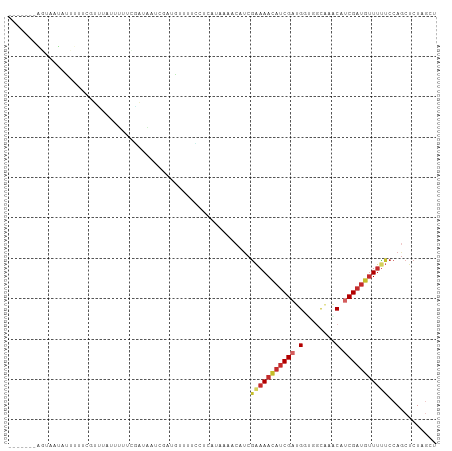
| Location | 5,859,910 – 5,860,012 |
|---|---|
| Length | 102 |
| Sequences | 9 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 60.56 |
| Shannon entropy | 0.84136 |
| G+C content | 0.37068 |
| Mean single sequence MFE | -22.03 |
| Consensus MFE | -7.63 |
| Energy contribution | -8.50 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.61 |
| SVM RNA-class probability | 0.999033 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5859910 102 - 22422827 ----AAGAGUAAUAUUUUUCGUUUAUUUUUCGAUAGUCAAUGUUCUCGCCAUAAAACAUCGAAAACAUCGACGGCGACGAGCAUCGAUGUUUUUCCAGCUCUAGCU ----.(((((........(((.........)))..(((.(((((((((((........((((.....)))).))))).)))))).))).........))))).... ( -23.20, z-score = -1.51, R) >droWil1.scaffold_181089 7826887 93 - 12369635 -----------AAACA-UCGAAACAUCGAUGUUUCAUCGAUGUUUGACGUUGAAAACAUCGAAAACAUCGAUGGUCCCAAACAUCGAUGUUUUUCCAGCUCUAGC- -----------(((((-((((..(((((((((((..(((((((((........))))))))))))))))))))((.....)).))))))))).............- ( -34.30, z-score = -5.59, R) >droSim1.chrX 4586294 102 - 17042790 ----AAGAGUAAUAUUUUUCGUUUAUUUUUCGAUAGUCAAUGUUCUCGCCAUAAAACAUCGAAAACAUCGACGGUGGAAAGCAUCGCUGUUUUUCCAGCUCUAGCU ----.(((((........(((((.((((((((((.((..(((.......)))...)))))))))).)).)))))(((((((((....)).)))))))))))).... ( -22.00, z-score = -1.49, R) >droSec1.super_4 5274257 100 + 6179234 ----AAGAGUAAUAUUUUUCGUUUA--UUUCGAUAGUCAAUGUUCUCCCCAUAAAACAUCGAAAACAUCGACGGCAGUAAGCAUCGAUGUUUUUCCAGCUCUAGCU ----.(((((...............--(((((((.((..(((.......)))...)))))))))(((((((..((.....)).))))))).......))))).... ( -20.70, z-score = -2.47, R) >droYak2.chrX 14713187 97 - 21770863 AAUUGUAAGUUCUUAUUUUGUU--UUUUUUCGACAAUCGAUUAUAGUAUC-------AUCGGAAACAUCGUUGUGGGCAGACAUCGACAUUUUACCAUCUCUUGCA ...((((((.........((((--.(((.((.(((((.(((......)))-------...(....)...))))).)).)))....))))...........)))))) ( -12.95, z-score = 0.80, R) >droEre2.scaffold_4690 3197778 91 - 18748788 -------GUUUGUUAUUCCGUUUCUUUUUUCGAUAAUCGAUUAUAGUCGC--------CCAAAAAUAUCGUUGCUGGCAAACAUCGAUGUUUUUCCAGCUCUAGCC -------....((((....((..((.(..(((.....)))..).))..))--------..............(((((.(((((....)))))..)))))..)))). ( -14.30, z-score = -0.82, R) >dp4.Unknown_group_23 12088 85 - 67933 -----------------GGCCGUUAAUGGGUUAAUUUCA----UUUGCUCUCAAAACAUCGAAAACAUCGAUGUUCGCAACCAUCGAUGUUUCUCCAGCUCUAGAU -----------------(((.((.((((((.....))))----)).))............(((((((((((((........))))))))))).))..)))...... ( -19.00, z-score = -1.79, R) >droPer1.super_0 2206017 99 - 11822988 -------AUAAGUUUAUAAGUUUUAUUUAAAAAACAUCGAUGUUUUGCUCCCAAAACAUCGUAAACAUCGAUGUCCGCAAACAUCGAUGUUUCUCCAGCUCUAUAU -------............(((((......)))))..((((((((((....)))))))))).((((((((((((......)))))))))))).............. ( -27.10, z-score = -6.90, R) >droMoj3.scaffold_6500 31529585 99 - 32352404 --AAUAGUUUUAAAUAUUCCGCGCAAC-AUGUAACAUCGAUGUUUUCUUCAAAACACAUCGAAAACAUCGAUGGUGGUAAACAUCGAUGAUAUUCCAGCUCU---- --....................((...-..(((.(((((((((((...(((.....((((((.....)))))).))).))))))))))).)))....))...---- ( -24.70, z-score = -3.41, R) >consensus _______AGUAAUAUUUUUCGUUUAUUUUUCGAUAAUCGAUGUUUUCCUCAUAAAACAUCGAAAACAUCGAUGGUGGCAAACAUCGAUGUUUUUCCAGCUCUAGCU ............................................................(((((((((((.(........).)))))))))))............ ( -7.63 = -8.50 + 0.87)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:19:14 2011