
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,810,717 – 5,810,906 |
| Length | 189 |
| Max. P | 0.898220 |

| Location | 5,810,717 – 5,810,831 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 64.66 |
| Shannon entropy | 0.64614 |
| G+C content | 0.44267 |
| Mean single sequence MFE | -22.80 |
| Consensus MFE | -8.54 |
| Energy contribution | -8.93 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.898220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
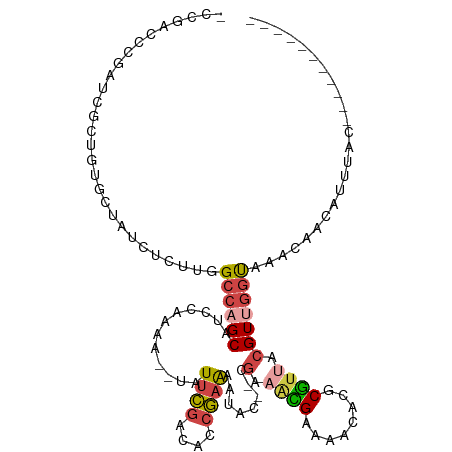
>dm3.chrX 5810717 114 + 22422827 UCCGACCCGAUCGCUGCGCUAUCUCUUGGCCAGCAUCCAAAA--UAUUCGACACCGAAAAUACAACGAAGCGAAAACACGCGUUACGUUGGUAAACAACAUUUACCAGGCUAACAG ..((....(((.((((.((((.....))))))))))).....--..((((....)))).......))..(((......)))((((..((((((((.....))))))))..)))).. ( -28.20, z-score = -2.74, R) >droSim1.chrX 4549889 103 + 17042790 UCCGACCCGAUCGCUGCGCUAUCUCUUGGCCAGCAUCCAAAA--UAUUCGGCACCGAAAAUACAACGAAACGAAAACACGCGUUACGUUGGUAAACAACAUUUAG----------- ........(((.((((.((((.....))))))))))).....--..((((....))))....(((((.((((........)))).)))))...............----------- ( -25.10, z-score = -2.41, R) >droSec1.super_4 5230624 103 - 6179234 UCCGACCCGAUCGCUGAGCUAUCUCUUGGCCAGCAUCCAAAA--UAUUCGGCACCGAAAAUACAACGAAACGAAAACACGCGUUACGUUGGUAAACAACAUUUAG----------- ........(((.((((.((((.....))))))))))).....--..((((....))))....(((((.((((........)))).)))))...............----------- ( -25.10, z-score = -2.68, R) >droYak2.chrX 14670663 95 + 21770863 ---------------GUGUUAUCUCUUGGCCAGCAUCCAAAA--UAUUCGACGCCGAAAAUAC---GAAAUGCAAACACGCGUUACGUCGGUAAACAACAUUUGCUAGCCGACUU- ---------------..........(((((.((((.......--...((((((.((......)---).(((((......))))).))))))...........)))).)))))...- ( -22.60, z-score = -1.60, R) >droEre2.scaffold_4690 3157596 91 + 18748788 ---------------GUGCUAUCUCUUGGCCAGCAUCCAAAA--UAUUCGACUCCGAAAAUAC---GAUGUGCAAACACGCGUUGCGUUGGUAAACAACGUUUGCCAACAU----- ---------------(.((((.....))))).(((.......--..((((....))))...((---(.((((....))))))))))((((((((((...))))))))))..----- ( -25.10, z-score = -2.23, R) >droAna3.scaffold_12929 113800 98 + 3277472 -UCCGCUGCCCGAAACUGUUAUCGCAUUGACAGCAUUCAAAAAUCAUUUAAAAAAACGAAUAU--UCUACUGCCGGUUCGCAUAACGUAAACAAAUUAGAA--------------- -..........(((.((((((......))))))..)))........................(--((((......(((..(.....)..)))....)))))--------------- ( -10.70, z-score = 0.89, R) >consensus _CCGACCCGAUCGCUGUGCUAUCUCUUGGCCAGCAUCCAAAA__UAUUCGACACCGAAAAUAC__CGAAACGAAAACACGCGUUACGUUGGUAAACAACAUUUAC___________ ............................((((((............((((....))))..........((((........))))..))))))........................ ( -8.54 = -8.93 + 0.39)


| Location | 5,810,791 – 5,810,906 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 69.74 |
| Shannon entropy | 0.57011 |
| G+C content | 0.49913 |
| Mean single sequence MFE | -35.68 |
| Consensus MFE | -17.90 |
| Energy contribution | -16.67 |
| Covariance contribution | -1.24 |
| Combinations/Pair | 1.65 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.746180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5810791 115 + 22422827 CACGCGUUACGUUGGUAAACAACAUUUACCAGGCUAACAGAUAUCAGCUGUUUGGGUAGUUGGAUCGCUGGAUUUAGAAGCUUUUGG-CGCCGUCUGGCAACGCUAGCGAUAGCUA ...(((((..((((.....)))).....((((((...((((((.....))))))(((.((..((..(((.........))).))..)-)))))))))).)))))((((....)))) ( -35.30, z-score = -1.09, R) >droSim1.chrX 4549963 104 + 17042790 CACGCGUUACGUUGGUAAACAACAUUUAG-----------AUAUCAGCUGUUUGGGUGGUUGGAUCGCUGGAUUUAGAAGCUUUCGG-UGCCGUCUGGCAACGCUGGUGGCAGCCA ...((.....((((.....)))).(((((-----------((.(((((.(((..(....)..))).)))))))))))).))....((-(((((.(..((...))..)))))).)). ( -33.30, z-score = -0.84, R) >droSec1.super_4 5230698 104 - 6179234 CACGCGUUACGUUGGUAAACAACAUUUAG-----------AUAUCAGCUGUUUGGGUGGUUGGAUCGCUAGAUUUAGGAGCUUUCGG-CACCGUCUGGCAACGCUGGUGGUAGCCA ...(((((..((((.....)))).....(-----------((.(((((..(....)..))))))))(((((((...((.((.....)-).)))))))))))))).(((....))). ( -33.50, z-score = -1.37, R) >droYak2.chrX 14670719 114 + 21770863 CACGCGUUACGUCGGUAAACAACAUUUGCUAG-CCGACUUAAAACAGCUGUUUGCGUGGUUGGAUCGCUGGAUUUAGGAGCUUUCGG-CGCCGUCUGGCAACGCUGGCGCUAAAAA ...((((((.((((((...((.....))...)-)))))...((((....))))((((.((..((((((((((..........)))))-))..)))..)).))))))))))...... ( -36.80, z-score = -0.30, R) >droEre2.scaffold_4690 3157652 110 + 18748788 CACGCGUUGCGUUGGUAAACAACGUUUGCCA-----ACAUUUAUCAGCUGUUUGUGUGGCUGGAUCGCUGGACUUAGGAGCUUUCGG-CGCCGUCUGGCAGCGCUGGCGGUGGCAA ......((((((((((((((...))))))))-----))........(((((..((((.((..((((((((((..........)))))-))..)))..)).))))..))))).)))) ( -43.50, z-score = -1.26, R) >droAna3.scaffold_12929 113873 101 + 3277472 UUCGCAUAACGUAAACAAAUUAGAAGGCACA-----------GCUGAUUGCCAGAGCUGCCAGAUGGCUAAAGGUUUAAGCUUUUUGUCACAAUAUGGCAGCUUUCGUGUCA---- .........................(((((.-----------((.....)).((((((((((..((((.((((((....)))))).)))).....)))))))))).))))).---- ( -31.70, z-score = -2.30, R) >consensus CACGCGUUACGUUGGUAAACAACAUUUACCA_________AUAUCAGCUGUUUGGGUGGUUGGAUCGCUGGAUUUAGAAGCUUUCGG_CGCCGUCUGGCAACGCUGGCGGUAGCAA ..........((((.....))))....................(((((((......)))))))((((((((.....(..(((..(((...)))...)))..).))))))))..... (-17.90 = -16.67 + -1.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:19:08 2011