
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,786,381 – 5,786,478 |
| Length | 97 |
| Max. P | 0.548380 |

| Location | 5,786,381 – 5,786,478 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 59.53 |
| Shannon entropy | 0.82193 |
| G+C content | 0.46522 |
| Mean single sequence MFE | -26.32 |
| Consensus MFE | -4.57 |
| Energy contribution | -5.28 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.88 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.17 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.548380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5786381 97 - 22422827 --CAGAAACGACGAUGAUGAACGUAGCUAUUGUAAUGACGAUAAUGGCCAUGAUCAUCGGAAACCGUU-UGGCGAUCGCUUGAAAAGCACAAGCACCUGC------ --((((((((.....(((((.(((.((((((((.......)))))))).))).)))))(....)))))-).......(((((.......)))))..))).------ ( -27.70, z-score = -2.64, R) >droPer1.super_21 1122741 87 + 1645244 --GCGAGAUGCUGCUAGUG--CUUGAUCAUCAUCAU--CAGCAGUAG-CAGCAGCACCAGAAACCGUU-GCUUGGGCG-----AGAGAACGAACUCCAGC------ --(.(((.((((((((.((--((((((....)))).--.)))).)))-)))))((.((((........-..)))))).-----..........))))...------ ( -28.90, z-score = -1.20, R) >droEre2.scaffold_4690 3133181 97 - 18748788 --CAGAAACAACGAUGAUGAACGUAGCUAUCGUAAUGACGAUAAUGGCCAUGAUCAUCGGAAACCGUU-UGGCGAUUGCUUGAAAAGCACAAGCACCUGC------ --......(((((..(((((.(((.((((((((....)))))...))).))).)))))(....))).)-))(((..((((((.......))))))..)))------ ( -26.30, z-score = -2.32, R) >droYak2.chrX 14645519 103 - 21770863 --CAGAAACGAUGAUGAUGAACGUAGCUAUCGUAAUGACGAUAAUGGCCAUGAUCAUCGGAAACCGUU-UGGCGAUUGCUUCAAAAGCACAAGCACAAGCACCUGC --(((...(((((((.(((..(((...((((((....)))))))))..))).)))))))......(((-((((...((((.....))))...)).)))))..))). ( -33.80, z-score = -3.94, R) >droSec1.super_4 5206459 97 + 6179234 --CAGAAACGACGAUUAUGAACGUAGCUAUCGUAAUGACGAUAAUGGCCAUGAUCAUCGGAAACCGUU-UGGCGAUCGCUUGAAAAGCACAAGCACCUGC------ --(((...(((.(((((((..(((...((((((....)))))))))..))))))).)))......(((-((((..((....))...)).)))))..))).------ ( -26.70, z-score = -2.11, R) >droSim1.chrX 4525430 97 - 17042790 --CAGAAACGACGAUGAUGAACGUAGCUAUCGUAAUGACGAUAAUGGCCAUGAUCAUCGGAAACCGUU-UGGCGAUCGCUUGAAAAGCACAAGCACCUGC------ --((((((((.....(((((.(((.((((((((....)))))...))).))).)))))(....)))))-).......(((((.......)))))..))).------ ( -26.50, z-score = -1.91, R) >droWil1.scaffold_180702 295929 91 + 4511350 -ACAAGAGCGAUAAUGGCCACCAC-ACAGCAAUACUCGUUGCGUUGG-AAUAAUUAUCUACGCCAUUUAACCUACUCGUUGGAUAAUCACAGGC------------ -........((((((..(((..((-.((((.......)))).)))))-....))))))...((((((((((......))))))).......)))------------ ( -16.21, z-score = -0.01, R) >droGri2.scaffold_14853 8101080 100 + 10151454 AUGUGUGUUAAUAAUUGAGCAUAUUUAACACACAAACACAUUAAAGCCCCGGCUUAUGUUAAACGAGCGAGCGAGUGAGCGAGAAGGCAAACGUUCCCGU------ .((((((((((.............))))))))))...((((..((((....))))))))...(((.(.(((((..((..(.....).))..)))))))))------ ( -24.42, z-score = -1.66, R) >consensus __CAGAAACGACGAUGAUGAACGUAGCUAUCGUAAUGACGAUAAUGGCCAUGAUCAUCGGAAACCGUU_UGGCGAUCGCUUGAAAAGCACAAGCACCUGC______ ........(((.(((.(((..(((....(((((....))))).)))..))).))).)))............................................... ( -4.57 = -5.28 + 0.70)

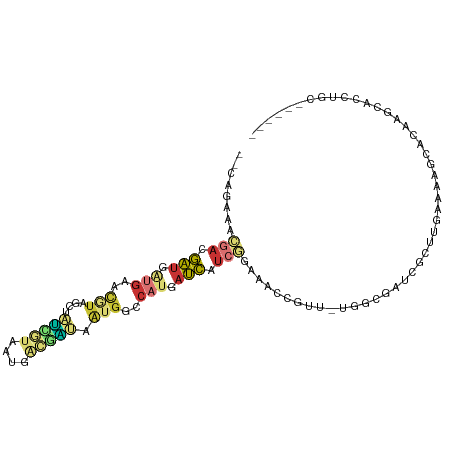
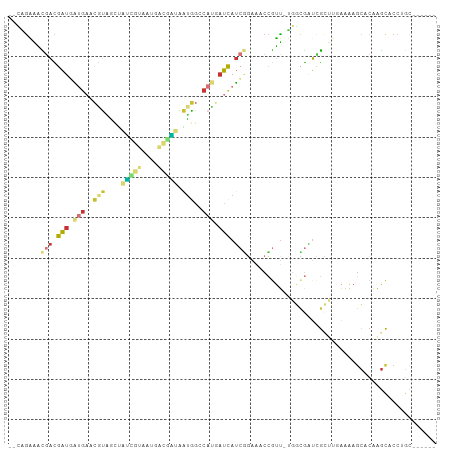
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:19:04 2011