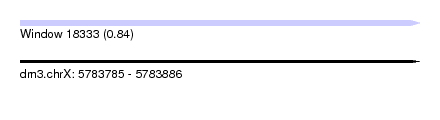
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,783,785 – 5,783,886 |
| Length | 101 |
| Max. P | 0.837008 |

| Location | 5,783,785 – 5,783,886 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 54.40 |
| Shannon entropy | 0.79483 |
| G+C content | 0.36683 |
| Mean single sequence MFE | -21.31 |
| Consensus MFE | -4.85 |
| Energy contribution | -4.27 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.23 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.837008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5783785 101 + 22422827 UGUACA-UACCCGCCAGUCUGCGCGCAGUGAAUUUCCCAAAUUAUU----UAUUCAUUCGCUGCCU-CUGUGAUCUCAUAGAUGAUUCAUUAAUUGAUGAUAUCAAU------ ......-....(((......))).(((((((((.............----.....))))))))).(-(((((....))))))(((((((((....))))).))))..------ ( -22.67, z-score = -2.11, R) >droSim1.chrX 4522964 98 + 17042790 ---------CAUACCCGCCUGCGAGCAGUGAAUUUCCCAAAUUAUU----UAUUCAUUCACUGCCU-CUGUGAUCUCAUAGAUGAUUCAUUAAUUGAUGAUAUCAAUAGCAG- ---------.........((((..(((((((((.............----.....))))))))).(-(((((....))))))(((((((((....))))).))))...))))- ( -25.87, z-score = -3.86, R) >droYak2.chrX 14642996 111 + 21770863 UAGACACUGUGCUCCCCCCGCUGAGCAGUGAAUUUCCCGAAUUAUUCAUUUAUUCAUUUACUGCCU-CUGUGAUCUCAUAGAUGAUUCAUUAAUGAAUUAUAUCAAUAGCAG- ...................((((.(((((((((.....((((.........))))))))))))).(-(((((....))))))(((((((....)))))))......))))..- ( -23.10, z-score = -2.34, R) >droAna3.scaffold_12929 83109 104 + 3277472 -----UUUUCAUAUUUGACUGCCUUUUCUGUGCUCUUAAAUGUGUU----CGUUCAUUAAUGGCUCGCUGUGAUCUCAUAGAUGAUUCACCAAUUGACGAUAUUAAUAAGUAG -----....(((((((((..(((......).))..))))))))).(----((((.(((..((..((((((((....))))).)))..))..))).)))))............. ( -23.00, z-score = -2.14, R) >dp4.chrXL_group1e 6918194 94 - 12523060 --------UUCCCUAUGACUAUCC-CAACAUGUUUCUACGGGUGUU---UCCCAAACUGUUUAUUAACAAUGAUUUCAUUGAUGAUUCACCAAUUAUUAGGAAUCC------- --------((((..((((......-..............(((....---.))).....((..((((.(((((....))))).))))..))...))))..))))...------- ( -15.30, z-score = -0.36, R) >droPer1.super_21 1119201 94 - 1645244 --------UUCCCCAUGACUAUCC-CAACAUUUUUCUACGGGUGUU---UCCCAAACUGUUUAUUAACAGUGAUUUCAUUGAUGAUUCACCAAUUAUUAGGAAUCC------- --------((((..((((..(((.-..............(((....---.)))..((((((....))))))))).))))((((((((....))))))))))))...------- ( -17.90, z-score = -1.34, R) >consensus ________UCCCACCUGACUGCCCGCAGUGAAUUUCCAAAAGUAUU____CAUUCAUUCACUGCCU_CUGUGAUCUCAUAGAUGAUUCACCAAUUAAUGAUAUCAAU______ ...................................................................(((((....)))))................................ ( -4.85 = -4.27 + -0.58)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:19:04 2011