| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,773,403 – 5,773,525 |
| Length | 122 |
| Max. P | 0.819323 |

| Location | 5,773,403 – 5,773,525 |
|---|---|
| Length | 122 |
| Sequences | 11 |
| Columns | 138 |
| Reading direction | reverse |
| Mean pairwise identity | 68.94 |
| Shannon entropy | 0.64218 |
| G+C content | 0.57054 |
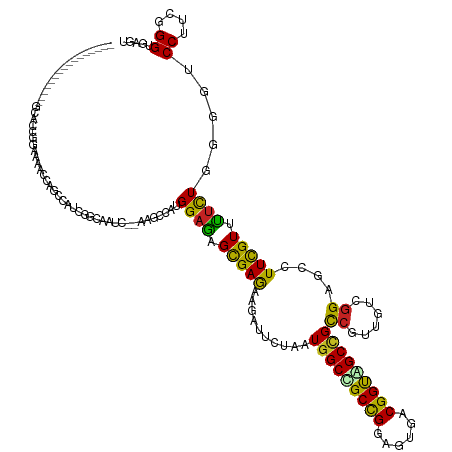
| Mean single sequence MFE | -43.92 |
| Consensus MFE | -23.43 |
| Energy contribution | -22.40 |
| Covariance contribution | -1.03 |
| Combinations/Pair | 1.55 |
| Mean z-score | -0.84 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.819323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
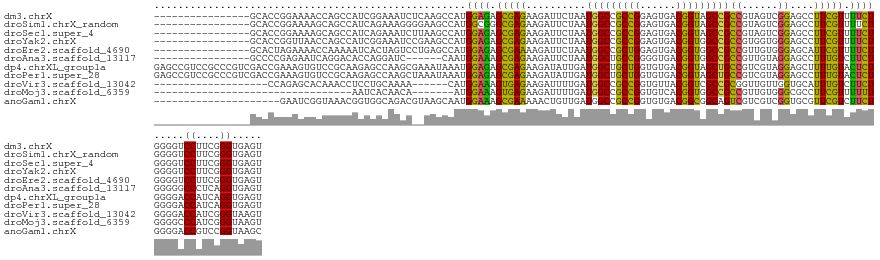
>dm3.chrX 5773403 122 - 22422827 ----------------GCACCGGAAAACCAGCCAUCGGAAAUCUCAAGCCAUGGAGAGCGAGAAGAUUCUAAUGGCCGCCGGAGUGACGGUAGCCGCCGUAGUCGGAGCCUUCGUUUUCUGGGGUCCUUCGGGUGAGU ----------------.((((.(((.(((.(((((..(((.((((..((........))..).))))))..)))))..((((((.(((((..((..(((....))).))..))))))))))))))..))).))))... ( -45.20, z-score = -0.78, R) >droSim1.chrX_random 1869190 122 - 5698898 ----------------GCACCGGAAAAGCAGCCAUCAGAAAGGGGAAGCCAUGGCGGGCGAGAAGAUUCUAAUGGCCGCCGGAGUGACGGUAGCCGCCGUAGUCGGAGCCUUCGUUUUCUGGGGUCCUUCGGGUGAGU ----------------.((((.(((.....(((.((((((((.(((.((..((((.(((.(((....)))...(((..(((......)))..))))))...))))..)).))).)))))))))))..))).))))... ( -49.20, z-score = -1.14, R) >droSec1.super_4 5194169 122 + 6179234 ----------------GCACCGGAAAAGCAGCCAUCAGAAAUCUUAAGCCAUGGAGAGCGAGAAGAUUCUAAUGGCCGCCGGAGUGACGGUAGCCGCCGUAGUCGGAGCCUUCGUUUUCUGGGGUCCUUCGGGUGAGU ----------------.((((.(((..((.(((((.((((.((((..((........))...)))))))).)))))..((((((.(((((..((..(((....))).))..))))))))))).))..))).))))... ( -44.30, z-score = -1.02, R) >droYak2.chrX 14632630 122 - 21770863 ----------------GCACCGGUUAACCAGCCAUCGGAAAUCCGAAGCCAUGGAGAGCGAGAAGAUUCUAAUGGCCGCCGGAGUGACGGUGGCCGCCGUGGUGGGAGCCUUCGUUUUCUGGGGUCCUUCGGGUGAGU ----------------.((((((((....)))).((((....)))).((((((((((((.....).))))...((((((((......)))))))).)))))))((((.(((.........))).))))...))))... ( -52.40, z-score = -1.75, R) >droEre2.scaffold_4690 3120563 122 - 18748788 ----------------GCACUAGAAAACCAAAAAUCACUAGUCCUGAGCCAUGGAGAGCGAAAAGAUUCUAAUGGCCGCUGGAGUGACGGUGGCCGCCGUUGUGGGAGCAUUCGUUUUCUGGGGUCCUUCGGGUGAGU ----------------....................(((...((((((((..((((((((((....(((((.(((((((((......)))))))))......)))))...))))))))))..)))...)))))..))) ( -41.30, z-score = -0.79, R) >droAna3.scaffold_13117 1201498 116 - 5790199 ----------------GCCCCGAGAAUCAGGACACCAGGAUC------CAAUGGAAAGCGAGAAGAUUCUAAUGGCUGCCGGGGUGACGGUGGCCGCCGUUGUAGGAGCCUUUGUCUUCUGGGGGCCCUCAGGUGAGU ----------------(((((.(((....(((((..(((.((------(.(..(...((.(((....)))...(((..(((......)))..)))))..)..).))).))).)))))))).))))).(((....))). ( -47.30, z-score = -1.17, R) >dp4.chrXL_group1a 7166516 138 + 9151740 GAGCCGUCCGCCCGUCGACCGAAAGUGUCCGCAAGAGCCAAGCGAAAUAAAUGGAGAGCGAGAAGAUAUUGAUGGCUGCUGGUGUGACGGUAGCUGCCGUCGUAGGAGCUUUUGUACUCUGGGGACCAUCAGGUGAGU ..(((.(((((((((.((((....).)))(((.........)))......))))...))).))......((((((.(.(..(.(((.(((.((((.((......)))))).)))))).)..).).))))))))).... ( -41.40, z-score = 0.54, R) >droPer1.super_28 681783 138 + 1111753 GAGCCGUCCGCCCGUCGACCGAAAGUGUCCGCAAGAGCCAAGCUAAAUAAAUGGAGAGCGAGAAGAUAUUGAUGGCUGCUGGUGUGACGGUAGCUGCCGUCGUAGGAGCCUUUGUACUCUGGGGACCAUCAGGUGAGU ..(((((((.((.((.((((....).))).))....................((((.(((((...........((((.(((...(((((((....)))))))))).))))))))).)))))))))).....))).... ( -40.30, z-score = 0.78, R) >droVir3.scaffold_13042 1797703 113 - 5191987 -------------------CCAGAGCACAAACCUCCUGCAAAA------CAUGGAAAGUGAGAAGAUUUUGAUGGCCGCCGGUGUUACGGUCGCCGCGGUUGUUGGUGCAUUUGUCUUCUGGGGACCAUCGGGUAAGU -------------------((((((.(((((.....((((...------(((.....)))........(..(..(((((.((((.......)))))))))..)..)))))))))).))))))..(((....))).... ( -35.60, z-score = -0.07, R) >droMoj3.scaffold_6359 1709704 98 - 4525533 ---------------------------------AAUCACAACA-------AUGGAAAGUGAGAAGAUUUUGAUGGCCGCCGGUGUCACGGUGGCCGCCGUUGUGGGCGCCUUCGUUUUUUGGGGCCCAUCGGGUAAGU ---------------------------------.(((...(((-------(((((((((......)))))...((((((((......)))))))).)))))))((((.((..........)).))))....))).... ( -39.20, z-score = -1.89, R) >anoGam1.chrX 4981143 117 - 22145176 ---------------------GAAUCGGUAAACGGUGGCAGACGUAAGCAAUGGAAAGCGAAAAACUGUUGAUGGCCGCCGGUGUGACGGCGGCAGUCGUCGUCGGUGCGUUCGUCUUCUGGGGACCGUCCGGUAAGC ---------------------..(((((...(((((.((........))...((((..((((..((((.((((((((((((......)))))))...))))).))))...))))..))))....)))))))))).... ( -46.90, z-score = -1.96, R) >consensus ________________GCACCGGAAAACCAGCCAUCGGCAAUC__AAGCCAUGGAGAGCGAGAAGAUUCUAAUGGCCGCCGGAGUGACGGUAGCCGCCGUUGUCGGAGCCUUCGUUUUCUGGGGUCCUUCGGGUGAGU ....................................................((((.(((((..........(((((((((......)))))))))((......))....))))).)))).....((....))..... (-23.43 = -22.40 + -1.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:19:02 2011