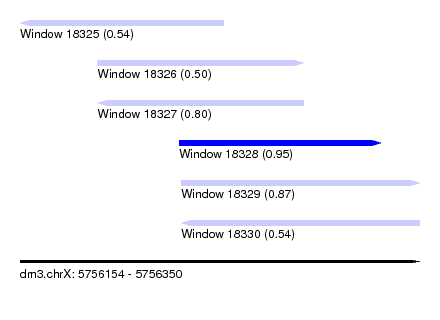
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,756,154 – 5,756,350 |
| Length | 196 |
| Max. P | 0.947493 |

| Location | 5,756,154 – 5,756,254 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 82.27 |
| Shannon entropy | 0.29580 |
| G+C content | 0.28831 |
| Mean single sequence MFE | -23.68 |
| Consensus MFE | -15.55 |
| Energy contribution | -15.80 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.540063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5756154 100 - 22422827 UUCUUAUUUCUGAUUUCAUUA------------------AUAAUUGAUCAGCGUGACAUUUCUGAUCGAUUCCUAUCAAAUUUGCAGCUUUUGUAAUUGGAAUUAAGUAUGCAACAAA ..((((.(((..(........------------------..((((((((((.(......).))))))))))..........((((((...)))))))..))).))))........... ( -16.80, z-score = -0.42, R) >droYak2.chrX 19053833 117 + 21770863 UUCCUAGUUCUGAAUUUAUUAGUAAUUGUUGAAAUAUUUAUAAUUGGUCAACGUGACAUUUAUGGCCGAUUUCGAUUAAAGUUGCAGA-UUUGCAAUUGGAAUUAAAUAUGCAACAAA ...(((((.........)))))...((((((..((((((..(((((((((............)))))))))..((((..((((((...-...))))))..)))))))))).)))))). ( -23.50, z-score = -1.24, R) >droSec1.super_4 5177598 117 + 6179234 UUCUUAUUUCUGAUUAUAUUAACAAUUUGUUAGGUAUUCAUAAUUGGUCAGCGUGACAUUUCUGAUCGAUUCCUAUUAAAGUUGCAGC-UUUGUAAUUGGAAUUAAGUAUGCAACAAA ..((((.(((..((((((....((((((..((((.......((((((((((.(......).))))))))))))))...))))))....-..))))))..))).))))........... ( -27.21, z-score = -2.59, R) >droSim1.chrX 4499947 117 - 17042790 UUCUUAUUUCUGAUUAUAUUAACAAUUUGUUAGGUAUUCAUAAUUGGUCAGCGUGACAUUUCUGAUCGAUUCCUAUUAAAGUUGCAGC-UUUGUAAUUGGAAUUAAGUAUGCAACAAA ..((((.(((..((((((....((((((..((((.......((((((((((.(......).))))))))))))))...))))))....-..))))))..))).))))........... ( -27.21, z-score = -2.59, R) >consensus UUCUUAUUUCUGAUUAUAUUAACAAUUUGUUAGGUAUUCAUAAUUGGUCAGCGUGACAUUUCUGAUCGAUUCCUAUUAAAGUUGCAGC_UUUGUAAUUGGAAUUAAGUAUGCAACAAA ..((((.((((..............................((((((((((.(......).))))))))))........((((((((...)))))))))))).))))........... (-15.55 = -15.80 + 0.25)

| Location | 5,756,192 – 5,756,293 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.93 |
| Shannon entropy | 0.35820 |
| G+C content | 0.34010 |
| Mean single sequence MFE | -22.25 |
| Consensus MFE | -11.05 |
| Energy contribution | -11.83 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chrX 5756192 101 + 22422827 UUUGAUAGGAAUCGAUCAGAAAUGUCACGCUGAUCAAUUAU------------------UAAUGAAAUCAGAAAUAAGAACUAGA-UUUCUUGCCAUACACAAAUACAUGGCUCGAAUCG (((((.(((((((((((((..........))))))..((((------------------(..((....))..)))))......))-))))).(((((..........))))))))))... ( -21.80, z-score = -2.21, R) >droYak2.chrX 19053870 118 - 21770863 UUUAAUCGAAAUCGGCCAUAAAUGUCACGUUGACCAAUUAUAAAUAUUUCAACAAUUACUAAUAAAUUCAGAACUAGGAACUGCACUUUCUGGCCGUACAUAA--ACAUGGCCGGAAUCG ......(((..((((((((..((((.(((((((...............)))))....................(((((((......)))))))..))))))..--..))))))))..))) ( -24.76, z-score = -2.73, R) >droSec1.super_4 5177635 119 - 6179234 UUUAAUAGGAAUCGAUCAGAAAUGUCACGCUGACCAAUUAUGAAUACCUAACAAAUUGUUAAUAUAAUCAGAAAUAAGAACUGGA-UUUCUGGCCAUACAUAAAUACAUGGCUCGAAUCG ........((.((((.((((((((((.....)))......(((.((..((((.....))))...)).)))..............)-))))))(((((..........))))))))).)). ( -20.20, z-score = -1.02, R) >consensus UUUAAUAGGAAUCGAUCAGAAAUGUCACGCUGACCAAUUAU_AAUA__U_A__AAUU__UAAUAAAAUCAGAAAUAAGAACUGGA_UUUCUGGCCAUACAUAAAUACAUGGCUCGAAUCG .............((((((..........))))))....................................................(((.((((((..........)))))).)))... (-11.05 = -11.83 + 0.78)


| Location | 5,756,192 – 5,756,293 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.93 |
| Shannon entropy | 0.35820 |
| G+C content | 0.34010 |
| Mean single sequence MFE | -26.24 |
| Consensus MFE | -16.45 |
| Energy contribution | -16.23 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.795629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5756192 101 - 22422827 CGAUUCGAGCCAUGUAUUUGUGUAUGGCAAGAAA-UCUAGUUCUUAUUUCUGAUUUCAUUA------------------AUAAUUGAUCAGCGUGACAUUUCUGAUCGAUUCCUAUCAAA .(((..((((((((........))))))..((((-((.((.........))))))))....------------------...(((((((((.(......).)))))))))))..)))... ( -23.10, z-score = -1.69, R) >droYak2.chrX 19053870 118 + 21770863 CGAUUCCGGCCAUGU--UUAUGUACGGCCAGAAAGUGCAGUUCCUAGUUCUGAAUUUAUUAGUAAUUGUUGAAAUAUUUAUAAUUGGUCAACGUGACAUUUAUGGCCGAUUUCGAUUAAA (((...((((((((.--((((((..((((((.....((((((.(((((.........))))).))))))..............)))))).))))))....))))))))...)))...... ( -30.11, z-score = -2.31, R) >droSec1.super_4 5177635 119 + 6179234 CGAUUCGAGCCAUGUAUUUAUGUAUGGCCAGAAA-UCCAGUUCUUAUUUCUGAUUAUAUUAACAAUUUGUUAGGUAUUCAUAAUUGGUCAGCGUGACAUUUCUGAUCGAUUCCUAUUAAA .((.((((((((((........))))))((((((-....(((..(((........)))..)))....(((((.((..(((....)))...)).))))))))))).)))).))........ ( -25.50, z-score = -1.49, R) >consensus CGAUUCGAGCCAUGUAUUUAUGUAUGGCCAGAAA_UCCAGUUCUUAUUUCUGAUUUUAUUA__AAUU__U_A__UAUU_AUAAUUGGUCAGCGUGACAUUUCUGAUCGAUUCCUAUUAAA ...(((..((((((........))))))..)))....(((.........))).............................((((((((((.(......).))))))))))......... (-16.45 = -16.23 + -0.22)

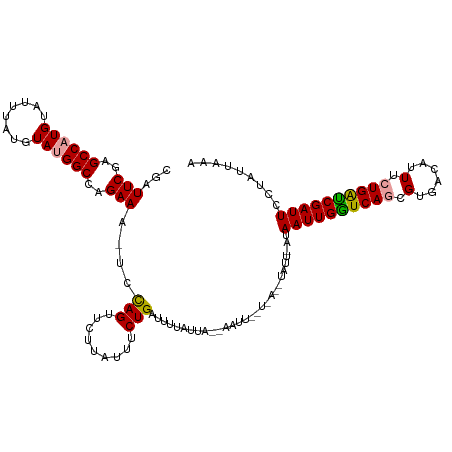
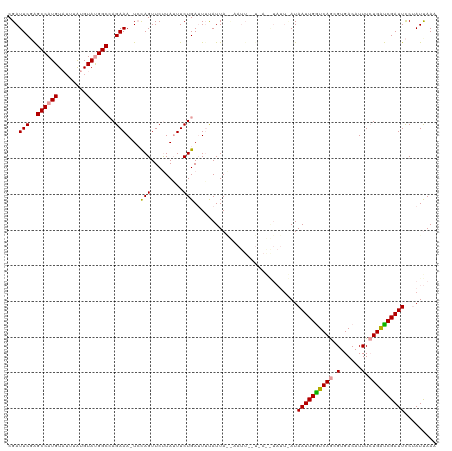
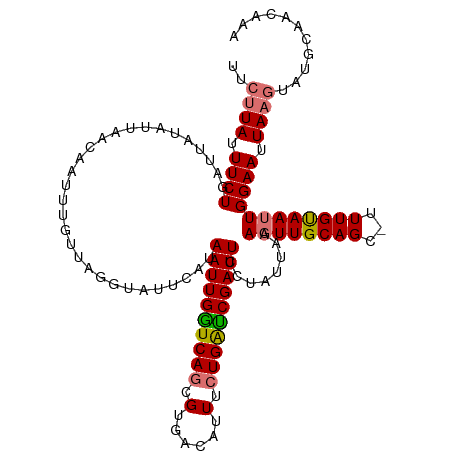
| Location | 5,756,232 – 5,756,331 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 71.71 |
| Shannon entropy | 0.39753 |
| G+C content | 0.41282 |
| Mean single sequence MFE | -26.13 |
| Consensus MFE | -18.02 |
| Energy contribution | -18.47 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5756232 99 + 22422827 ------------------UUAAUGAAAUCAGAAAUAAGAACUAGA-UUUCUUGCCAUACACAAAUACAUGGCUCGAAUCGAUAUCUUGGCCCAAAGGCCAUCGAUCUUGGUCCGGGCC ------------------.....((((((((.........)).))-))))...................((((((.(((((.(((.(((((....)))))..))).))))).)))))) ( -28.00, z-score = -2.73, R) >droYak2.chrX 19053910 112 - 21770863 UAAAUAUUUCAACAAUUACUAAUAAAUUCAGAACUAGGAACUGCACUUUCUGGCCGUACAUAAA--CAUGGCCGGAAUCGAUUUCGUGGCCCAAGAACCAUCGAUCUUGGCCUG---- ....................................((((.((....((((((((((.......--.)))))))))).)).))))..((((.((((........))))))))..---- ( -25.90, z-score = -2.11, R) >droSec1.super_4 5177675 114 - 6179234 UGAAUACCUAACAAAUUGUUAAUAUAAUCAGAAAUAAGAACUGGA-UUUCUGGCCAUACAUAAAUACAUGGCUCGAAUCGAUUCCUUGGCCCAAAGGCCAUCGAUCUUGGCCCGG--- (((.((..((((.....))))...)).)))..........((((.-.(((.((((((..........)))))).)))..((((...(((((....)))))..)))).....))))--- ( -24.50, z-score = -0.80, R) >consensus U_AAUA__U_A__AAUU_UUAAUAAAAUCAGAAAUAAGAACUGGA_UUUCUGGCCAUACAUAAAUACAUGGCUCGAAUCGAUUUCUUGGCCCAAAGGCCAUCGAUCUUGGCCCGG___ ..................................(((((........(((.((((((..........)))))).)))((((.....(((((....))))))))))))))......... (-18.02 = -18.47 + 0.45)

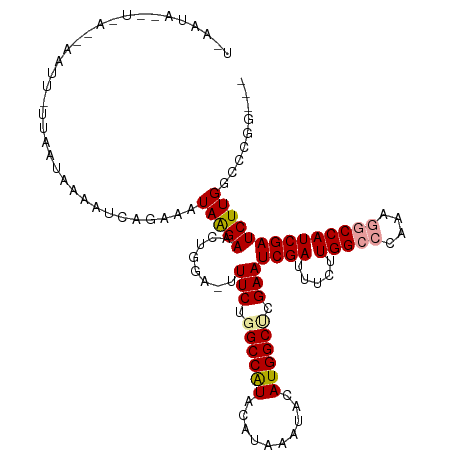
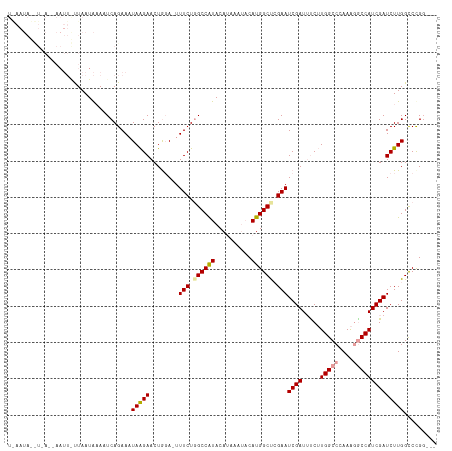
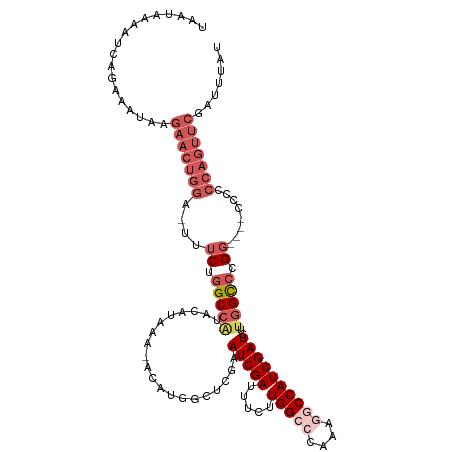
| Location | 5,756,233 – 5,756,350 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 78.29 |
| Shannon entropy | 0.37497 |
| G+C content | 0.45050 |
| Mean single sequence MFE | -30.92 |
| Consensus MFE | -14.83 |
| Energy contribution | -17.90 |
| Covariance contribution | 3.07 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.873189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5756233 117 + 22422827 UAAUGAAAUCAGAAAUAAGAACUAGA-UUUCUUGCCAUACACAAAUACAUGGCUCGAAUCGAUAUCUUGGCCCAAAGGCCAUCGAUCUUGGUCCGGGCCCCCCCCCAGUUCGAUUUAU .....((((((....)..(((((.((-(((...(((((..........)))))..)))))........(((((...(((((.......))))).))))).......)))))))))).. ( -31.70, z-score = -2.27, R) >droEre2.scaffold_4690 3102354 111 + 18748788 UAAUAAAACCAGAACCAAUUUCUGGACUUUCUGGCCAUACAUAAA-AGAUGG-CCAAAUCGAUUUCAUGGCCCAAACACCAUCGAUCUUGGCCUGC-----CACCCAGUUCGAUUUAU ........((((((.....)))))).......((((((.(.....-.)))))-))(((((((.....((((((((............))))...))-----))......))))))).. ( -29.10, z-score = -3.35, R) >droYak2.chrX 19053929 111 - 21770863 UAAUAAAUUCAGAACUAGGAACUGCACUUUCUGGCCGUACAUAAA--CAUGGCCGGAAUCGAUUUCGUGGCCCAAGAACCAUCGAUCUUGGCCUGG-----CCCCCCAUUCGAUUUAU ..(((((((..((((((((.....(((.((((((((((.......--.))))))))))..(....))))..((((((........)))))))))))-----.......)))))))))) ( -28.51, z-score = -1.13, R) >droSec1.super_4 5177694 113 - 6179234 UAAUAUAAUCAGAAAUAAGAACUGGA-UUUCUGGCCAUACAUAAAUACAUGGCUCGAAUCGAUUCCUUGGCCCAAAGGCCAUCGAUCUUGGCCCGG----CCCCCCAGUUCGAUUUAU ..................(((((((.-.(((.((((((..........)))))).)))..........((((....(((((.......))))).))----))..)))))))....... ( -34.80, z-score = -2.95, R) >droSim1.chrX 4500043 95 + 17042790 UAAUAUAAUCAGAAAUAAGAACUGGA-UUUCUGGC------------------UCGAAUCGAUUCCUUGGCCCACAGGCCAUCGAUCUUGGCCCGG----CCCCCCAGUUCGAUUUAU ..................(((((((.-...((((.------------------.((((((((.....(((((....)))))))))).)))..))))----....)))))))....... ( -30.50, z-score = -2.67, R) >consensus UAAUAAAAUCAGAAAUAAGAACUGGA_UUUCUGGCCAUACAUAAA_ACAUGGCUCGAAUCGAUUUCUUGGCCCAAAGGCCAUCGAUCUUGGCCCGG____CCCCCCAGUUCGAUUUAU ..................(((((((...(((.((((((..........)))))).)))((((.....(((((....)))))))))...................)))))))....... (-14.83 = -17.90 + 3.07)

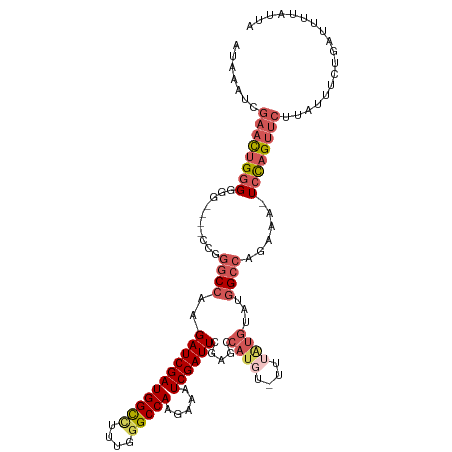
| Location | 5,756,233 – 5,756,350 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.29 |
| Shannon entropy | 0.37497 |
| G+C content | 0.45050 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -21.90 |
| Energy contribution | -22.98 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.544900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5756233 117 - 22422827 AUAAAUCGAACUGGGGGGGGGCCCGGACCAAGAUCGAUGGCCUUUGGGCCAAGAUAUCGAUUCGAGCCAUGUAUUUGUGUAUGGCAAGAAA-UCUAGUUCUUAUUUCUGAUUUCAUUA ...((((((.(((((......)))))......(((..(((((....))))).))).))))))...((((((........))))))..((((-((.((.........)))))))).... ( -36.10, z-score = -1.07, R) >droEre2.scaffold_4690 3102354 111 - 18748788 AUAAAUCGAACUGGGUG-----GCAGGCCAAGAUCGAUGGUGUUUGGGCCAUGAAAUCGAUUUGG-CCAUCU-UUUAUGUAUGGCCAGAAAGUCCAGAAAUUGGUUCUGGUUUUAUUA ...(((.((((..(.((-----(....))).(((((((....(((((((..((....)).(((((-(((((.-.....).)))))))))..))))))).))))))))..)))).))). ( -33.50, z-score = -1.51, R) >droYak2.chrX 19053929 111 + 21770863 AUAAAUCGAAUGGGGGG-----CCAGGCCAAGAUCGAUGGUUCUUGGGCCACGAAAUCGAUUCCGGCCAUG--UUUAUGUACGGCCAGAAAGUGCAGUUCCUAGUUCUGAAUUUAUUA (((((((((((.(((((-----(..((((..(((((((((((....))))).....))))))..))))...--....(((((.........))))))))))).)))).).)))))).. ( -34.70, z-score = -1.16, R) >droSec1.super_4 5177694 113 + 6179234 AUAAAUCGAACUGGGGGG----CCGGGCCAAGAUCGAUGGCCUUUGGGCCAAGGAAUCGAUUCGAGCCAUGUAUUUAUGUAUGGCCAGAAA-UCCAGUUCUUAUUUCUGAUUAUAUUA .......((((((((.((----((.(((...(((((((((((....))))).....))))))...)))..((((....)))))))).....-)))))))).................. ( -38.90, z-score = -2.18, R) >droSim1.chrX 4500043 95 - 17042790 AUAAAUCGAACUGGGGGG----CCGGGCCAAGAUCGAUGGCCUGUGGGCCAAGGAAUCGAUUCGA------------------GCCAGAAA-UCCAGUUCUUAUUUCUGAUUAUAUUA .......((((((((...----...(((...(((((((((((....))))).....))))))...------------------))).....-)))))))).................. ( -31.30, z-score = -1.30, R) >consensus AUAAAUCGAACUGGGGGG____CCGGGCCAAGAUCGAUGGCCUUUGGGCCAAGAAAUCGAUUCGAGCCAUGU_UUUAUGUAUGGCCAGAAA_UCCAGUUCUUAUUUCUGAUUUUAUUA .......(((((((((......)).((((..(((((((((((....))))).....)))))).....((((....))))...)))).......))))))).................. (-21.90 = -22.98 + 1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:19:01 2011