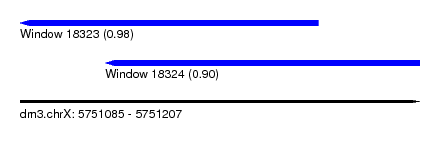
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,751,085 – 5,751,207 |
| Length | 122 |
| Max. P | 0.977267 |

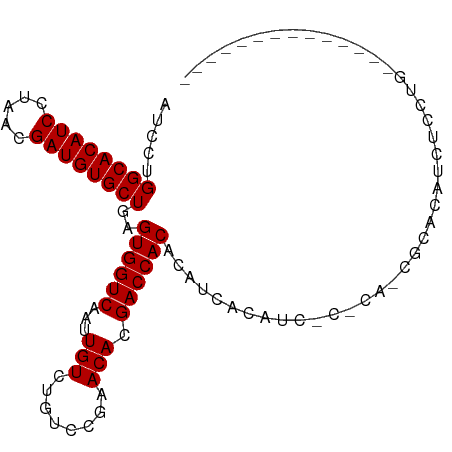
| Location | 5,751,085 – 5,751,176 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 81.20 |
| Shannon entropy | 0.30502 |
| G+C content | 0.51632 |
| Mean single sequence MFE | -22.76 |
| Consensus MFE | -21.06 |
| Energy contribution | -21.06 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.977267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5751085 91 - 22422827 AUCCUGGCACAUCCUAACGAUGUGCUGAGUGGUCAAUUGUCUGUCCGAACACGACCACACAUCACAUCGCACAUCGCACAUCUCACAUCUC---------- .....((((((((.....))))))))..((((((.....((.....))....))))))..........((.....))..............---------- ( -22.20, z-score = -1.41, R) >droSec1.super_4 5165554 80 + 6179234 AUCCUGGCACAUCCUAACGAUGUGCUGAGUGGUCAAUUGUGUGUCCGAACACGACCACACAUCACAU-------CGCACAUCUCCUG-------------- .....((((((((.....))))))))..(((((....((((((..((....))..))))))....))-------)))..........-------------- ( -25.40, z-score = -2.39, R) >droSim1.chrX 4493609 75 - 17042790 AUCCUGGCACAUCCUAACGAUGUGCUGAGUGGUCAAUUGUCUGUCCGAACACGACCACAC-----AU-------CGCACAUCUCCUG-------------- ..................(((((((((.((((((.....((.....))....)))))).)-----).-------.))))))).....-------------- ( -21.10, z-score = -1.91, R) >droYak2.chrX_random 211730 87 - 1802292 AUCCUGGCACAUCCUAACGAUGUGCUGAGUGGUCAAUUGUCUGUCCGAACACGACCACACAUCACAUCUCGCACCGCACAUCUCCUG-------------- .....((((((((.....))))))))..((((((.....((.....))....)))))).............................-------------- ( -20.90, z-score = -1.16, R) >droEre2.scaffold_4690 3097393 101 - 18748788 AUCCUGGCACAUCCUAACGAUGUGCUGAGUGGUCAAUUGUCUGUCCGCACAUGACCACACAUCACCACUCACAUCUCACAUCUCACAUCCCACGUCUCCUG .....((((((((.....))))))))(((((((....(((..(((.......)))...)))..)))))))............................... ( -24.20, z-score = -2.23, R) >consensus AUCCUGGCACAUCCUAACGAUGUGCUGAGUGGUCAAUUGUCUGUCCGAACACGACCACACAUCACAUC_C_CA_CGCACAUCUCCUG______________ .....((((((((.....))))))))..((((((...(((........))).))))))........................................... (-21.06 = -21.06 + 0.00)

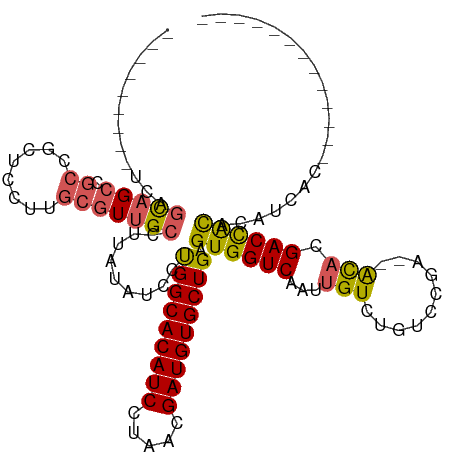
| Location | 5,751,111 – 5,751,207 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.56 |
| Shannon entropy | 0.35914 |
| G+C content | 0.54794 |
| Mean single sequence MFE | -31.54 |
| Consensus MFE | -21.19 |
| Energy contribution | -21.39 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.904038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5751111 96 - 22422827 ---------UCAGCAGCCGCCGCUCCUUGCGUUGCCUUAUAUCCUGGCACAUCCUAACGAUGUGCUGAGUGGUCAAUUGUCUGUCCGA--ACACGACCACACAUCAC------------- ---------...(((((.((........)))))))..........((((((((.....))))))))..((((((.....((.....))--....)))))).......------------- ( -27.70, z-score = -1.74, R) >droSec1.super_4 5165569 96 + 6179234 ---------UCAGCAGCCGCCGCUCCUUGCGUUGCCUUAUAUCCUGGCACAUCCUAACGAUGUGCUGAGUGGUCAAUUGUGUGUCCGA--ACACGACCACACAUCAC------------- ---------...(((((.((........)))))))..........((((((((.....))))))))..((((((....((((......--)))))))))).......------------- ( -30.90, z-score = -2.28, R) >droSim1.chrX 4493621 94 - 17042790 ---------UCAGCAGCCGCCGCUCCUUGCGUUGCCUUAUAUCCUGGCACAUCCUAACGAUGUGCUGAGUGGUCAAUUGUCUGUCCGA--ACACGACCACACAUC--------------- ---------...(((((.((........)))))))..........((((((((.....))))))))..((((((.....((.....))--....)))))).....--------------- ( -27.70, z-score = -1.82, R) >droEre2.scaffold_4690 3097429 105 - 18748788 GAUUGGUAUUCAGCAGCCGCCGCUCCUUGCGUUGCCUUAUAUCCUGGCACAUCCUAACGAUGUGCUGAGUGGUCAAUUGUCUGUCCGC--ACAUGACCACACAUCAC------------- (((.(((((...(((((.((........)))))))...)))))..((((((((.....))))))))..(((((((..(((......))--)..)))))))..)))..------------- ( -33.50, z-score = -2.12, R) >droAna3.scaffold_13117 1180626 100 - 5790199 ---------UCAGUAG----AGCUCCUUGGCUCG-------UCCCGGCACAUCCUAACGAUGUGCUGAGUGGUCAGUUGUCUGUCCGGCUGUCCGACUGUCCGAGUGUCCGACGACUCAC ---------......(----((.((.((((((((-------..((((((((((.....)))))))))(((((.((((((......)))))).)).))))..))))...)))).))))).. ( -37.90, z-score = -1.68, R) >consensus _________UCAGCAGCCGCCGCUCCUUGCGUUGCCUUAUAUCCUGGCACAUCCUAACGAUGUGCUGAGUGGUCAAUUGUCUGUCCGA__ACACGACCACACAUCAC_____________ ............(((((.((........))))))).........(((((((((.....))))))))).((((((....................)))))).................... (-21.19 = -21.39 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:18:56 2011