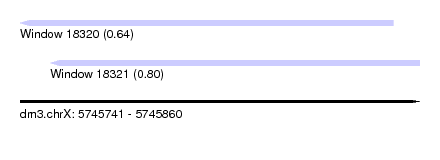
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,745,741 – 5,745,860 |
| Length | 119 |
| Max. P | 0.795480 |

| Location | 5,745,741 – 5,745,852 |
|---|---|
| Length | 111 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 74.87 |
| Shannon entropy | 0.50141 |
| G+C content | 0.36376 |
| Mean single sequence MFE | -13.80 |
| Consensus MFE | -6.82 |
| Energy contribution | -6.69 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.641571 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5745741 111 - 22422827 GCUUGUGUUUAUGCAAAUAUUUGUAUUCCAUUUGCAUUUCAAUAAUUGAAA--UUUUUAUUUUUUCUGCCUUUGCGCUCUUGUUUUUCUCGCACCCGCGCCCGCGCCAUUCCG ((..(((...((((((....))))))..)))..))((((((.....)))))--).............((....((((...(((.......)))...))))..))......... ( -17.70, z-score = -1.28, R) >droPer1.super_28 484792 110 - 1111753 GCCUGUGUUUAUGCAAAUAUUUGUAUUCCAUUUGCAUUUCAAUAAUUGAAA--UUUUUAUUUUCGUUUCUCGCUUGCCCUUACUUCUUUACUUCCACUAUCUCUCUCACUCG- ((..(((...((((((((...........))))))))..........((((--.......))))......)))..))...................................- ( -9.90, z-score = -2.13, R) >dp4.chrXL_group1a 2050211 110 - 9151740 GCCUGUGUUUAUGCAAAUAUUUGUAUUCCAUUUGCAUUUCAAUAAUUGAAA--UUUUUAUUUUCGUUUCUCGCUUGCCCUUACUUCUUUACUUCCACUAUCUCUCUCACUCG- ((..(((...((((((((...........))))))))..........((((--.......))))......)))..))...................................- ( -9.90, z-score = -2.13, R) >droSim1.chrX 4487966 105 - 17042790 GCUUGUGUUUAUGCAAAUAUUUGUAUUCCAUUUGCAUUUCAAUAAUUGAAA--UUUUUAUUUUUUCUGCCUUUGCGCUCUUGUUCUUC------CCGCACCCCCGCCAUUCCG ((..(((...((((((....))))))..)))..))((((((.....)))))--)...................(((....(((.....------..)))....)))....... ( -12.70, z-score = -1.44, R) >droSec1.super_4 5159941 105 + 6179234 GCUUGUGUUUAUGCAAAUAUUUGUAUUCCAUUUGCAUUUCAAUAAUUGAAA--UUUUUUUUUUUUCUGCCUUUGCGCUCUUGUUUUUC------CCGCACCCCCGCCAUUCCG ((..(((...((((((....))))))..)))..))((((((.....)))))--)...................(((....(((.....------..)))....)))....... ( -12.70, z-score = -1.41, R) >droYak2.chrX 19046944 110 + 21770863 GCUUGUGUUUAUGCAAAUAUUUGUAUUCCAUUUGCAUUUCAAUAAUUGAAA---UUUUAUUUUUUCUGCCUUGGCGCUCUUGUUUUUUCCGCUCCCUCGCCAUUGCCAUUCCG ((..(((...((((((....))))))..)))..))((((((.....)))))---)............((..(((((.....((.......)).....)))))..))....... ( -19.60, z-score = -2.64, R) >droEre2.scaffold_4690 3093042 108 - 18748788 GCUUGUGUUUAUGCAAAUAUUUGUAUUCCAUUUGCAUUUCAAUAAUUGAAAAUUUUUUGUUUUUUCCGCCCUUGCGCUCUUGUUUUUC-----GCCGACCCGUCGCCAUUCCG ((..(((...((((((....))))))..)))..)).(((((.....)))))...............(((....)))............-----(.(((....))).)...... ( -13.90, z-score = -0.49, R) >droWil1.scaffold_181096 1847579 104 - 12416693 GCUUGUGUUUGAAGCAAAAAUUGUGUUUGAUAAGUAUUUGAGAAAUUUAUG--UUGUUAUUUUUGCUGGUUUUGCGCUAUUUUCCUUC-----UCGUCAUCCUCCUUUUUC-- ....(((...((((..(((((.((((..(((.((((..(((.((.......--)).)))....)))).)))..)))).))))).))))-----....)))...........-- ( -14.00, z-score = -0.11, R) >consensus GCUUGUGUUUAUGCAAAUAUUUGUAUUCCAUUUGCAUUUCAAUAAUUGAAA__UUUUUAUUUUUUCUGCCUUUGCGCUCUUGUUUUUC_____CCCGCACCCCCGCCAUUCCG ((..(((...((((((....))))))..)))..)).((((((...)))))).............................................................. ( -6.82 = -6.69 + -0.14)
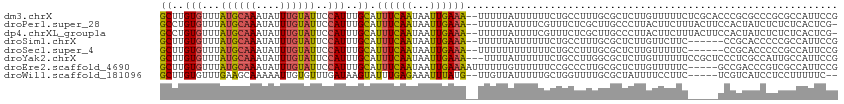
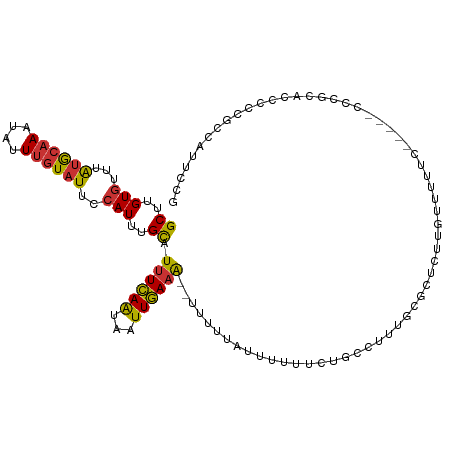
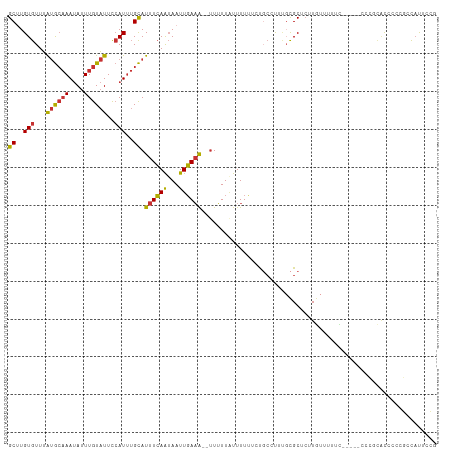
| Location | 5,745,750 – 5,745,860 |
|---|---|
| Length | 110 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 68.78 |
| Shannon entropy | 0.64154 |
| G+C content | 0.36241 |
| Mean single sequence MFE | -14.85 |
| Consensus MFE | -6.29 |
| Energy contribution | -6.28 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.795480 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 5745750 110 - 22422827 UGCGCUUCGCUUGUGUUUAUGCAAAUAUUUGUAUUCCAUUUGCAUUUCAAUAAUUGAAA--UUUUUAUUUUUUCUGCC-UUUGCGCUCUUGUUUUUCUCGCACCCGCGCCCGC .((((...((..(((...((((((....))))))..)))..))((((((.....)))))--)................-...))))............(((....)))..... ( -19.80, z-score = -1.38, R) >droAna3.scaffold_13117 4612131 84 + 5790199 --CGCUCCGCCAGUGUUUAUGCAAAUAUUUGUAUUCCAUUUGCAUUCCAAUAAUUGAAA--UUUUUACCCUCCCAGCUCCGCCGGGUC------------------------- --......((.((((...((((((....))))))..)))).))................--..........(((.((...)).)))..------------------------- ( -13.00, z-score = -0.55, R) >droPer1.super_28 484801 109 - 1111753 UUCGCAUUGCCUGUGUUUAUGCAAAUAUUUGUAUUCCAUUUGCAUUUCAAUAAUUGAAA--UUUUUAUUUUCGUUUCU-CGCUUGCCCUUACUUCUUUACUUCCACUAUCUC- ...(((..((...((...((((((((...........))))))))..))......((((--.......))))......-.)).)))..........................- ( -11.90, z-score = -1.79, R) >dp4.chrXL_group1a 2050220 109 - 9151740 UUCGCAUUGCCUGUGUUUAUGCAAAUAUUUGUAUUCCAUUUGCAUUUCAAUAAUUGAAA--UUUUUAUUUUCGUUUCU-CGCUUGCCCUUACUUCUUUACUUCCACUAUCUC- ...(((..((...((...((((((((...........))))))))..))......((((--.......))))......-.)).)))..........................- ( -11.90, z-score = -1.79, R) >droSim1.chrX 4487975 104 - 17042790 UGCGCUUCGCUUGUGUUUAUGCAAAUAUUUGUAUUCCAUUUGCAUUUCAAUAAUUGAAA--UUUUUAUUUUUUCUGCC-UUUGCGCUCUUGUUCUUCCCGCACCCCC------ .((((...((..(((...((((((....))))))..)))..))((((((.....)))))--)................-...)))).....................------ ( -17.60, z-score = -2.78, R) >droYak2.chrX 19046953 107 + 21770863 --CGCUUCGCUUGUGUUUAUGCAAAUAUUUGUAUUCCAUUUGCAUUUCAAUAAUUGAAA---UUUUAUUUUUUCUGCC-UUGGCGCUCUUGUUUUUUCCGCUCCCUCGCCAUU --.((...((..(((...((((((....))))))..)))..))((((((.....)))))---)............)).-.(((((.....((.......)).....))))).. ( -18.10, z-score = -1.98, R) >droEre2.scaffold_4690 3093051 103 - 18748788 ----CUUCGCUUGUGUUUAUGCAAAUAUUUGUAUUCCAUUUGCAUUUCAAUAAUUGAAAAUUUUUUGUUUUUUCCGCC-CUUGCGCUCUUGUUUUUCGCCGACCCGUC----- ----....((..(((...((((((....))))))..)))..)).(((((.....)))))...............(((.-...))).......................----- ( -11.90, z-score = -0.29, R) >droWil1.scaffold_181096 1847587 104 - 12416693 CUUGUUUUGCUUGUGUUUGAAGCAAAAAUUGUGUUUGAUAAGUAUUUGAGAAAUUUAUG--UUGUUAUUUUUGCUGGU-UUUGCGCUAUUUUCCUUCUCGUCAUCCU------ ....((((((((.......))))))))...((((..(((.((((..(((.((.......--)).)))....)))).))-)..)))).....................------ ( -14.60, z-score = -0.07, R) >consensus U_CGCUUCGCUUGUGUUUAUGCAAAUAUUUGUAUUCCAUUUGCAUUUCAAUAAUUGAAA__UUUUUAUUUUUUCUGCC_CUUGCGCUCUUGUUUUUCUCGCACCCCU______ ........((..(((...((((((....))))))..)))..)).((((((...))))))...................................................... ( -6.29 = -6.28 + -0.01)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:18:54 2011