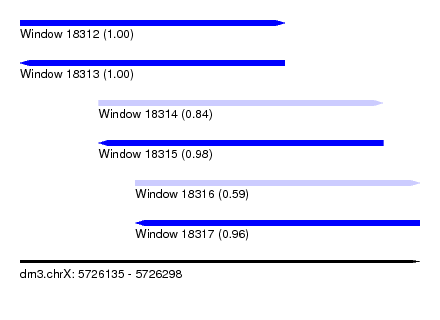
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,726,135 – 5,726,298 |
| Length | 163 |
| Max. P | 0.999191 |

| Location | 5,726,135 – 5,726,243 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.00 |
| Shannon entropy | 0.43526 |
| G+C content | 0.59720 |
| Mean single sequence MFE | -44.84 |
| Consensus MFE | -36.42 |
| Energy contribution | -37.46 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.70 |
| SVM RNA-class probability | 0.999191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5726135 108 + 22422827 --------UUUCCAUUGCUACUUUUGAUAACUUCACCGGGAUUCGAACCCGGACCGGCGGCACUACCUCCCG---AUUGGAAGUUCGCAUACUAACUGUAUGCGCUGCCGCCGGCGGCG --------........(((................(((((.......))))).(((((((((((.((.....---...)).))).(((((((.....)))))))..)))))))).))). ( -43.90, z-score = -4.59, R) >droSim1.chrX 4465015 109 + 17042790 --------UUUCCGUUGCUACUUUUAUUAACUUCACCGGGAUUCGAACCCGGACCGGCGGCACCACACCCGAG--AUCGGGAGUUCGCAUACUAGCUGUAUGCGCUGCCGCCGGCGGCG --------....((((((.................(((((.......)))))..((((((((..((.((((..--..)))).)).(((((((.....))))))).)))))))))))))) ( -50.20, z-score = -4.73, R) >droSec1.super_4 5139713 108 - 6179234 --------UUUCCGUUGCUACUUUUAUUAACUUCACCGGGAUUCGAACCCGGACCGGCGGCACCACACCCGA---AUCGGGAGUUCGCAUACUAGCUGUAUGCGCUGCCGCCGGCGGCG --------....((((((.................(((((.......)))))..((((((((..((.((((.---..)))).)).(((((((.....))))))).)))))))))))))) ( -48.00, z-score = -4.32, R) >droYak2.chrX 19025693 113 - 21770863 UGUAUCCAU-UUCCAUGCUACUUUA-----CUUCACCGGGAUUCGAACCCGGACCGGCGGCACUACUCCGCUCCGUUUGGGAGCUCGGAGUCCAACCAACUGUGCUGCCGCCGGCGGCG .........-......(((......-----.....(((((.......))))).(((((((((..((((((((((.....)))))..))))).((......))...))))))))).))). ( -44.50, z-score = -2.63, R) >droEre2.scaffold_4690 3074836 111 + 18748788 CGUGUCUGUGUUCCAUGCUACUUUUAC---UUUCACCGGGACUCGAACCCGGACCGGCGGCUCUACUCCG-----UGUGGGCAGUCGGAGCCUAACUACCUGGACUACCGCCGGCGGCG ................(((........---.....(((((.......))))).((((((((((((((((.-----....)).))).)))))).........((....))))))).))). ( -37.60, z-score = 0.13, R) >consensus ________UUUCCCUUGCUACUUUUAUUAACUUCACCGGGAUUCGAACCCGGACCGGCGGCACUACACCCCA___AUUGGGAGUUCGCAUACUAACUGUAUGCGCUGCCGCCGGCGGCG ................(((................(((((.......))))).(((((((((.....(((........)))....(((((((.....))))))).))))))))).))). (-36.42 = -37.46 + 1.04)

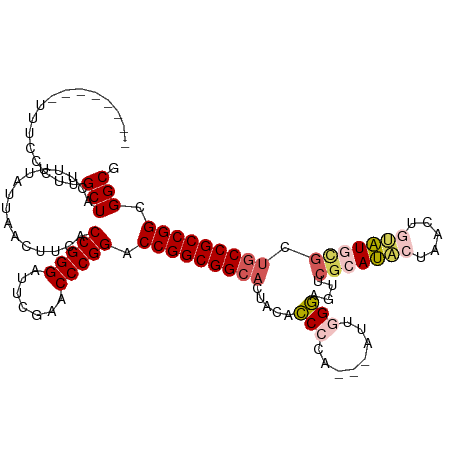
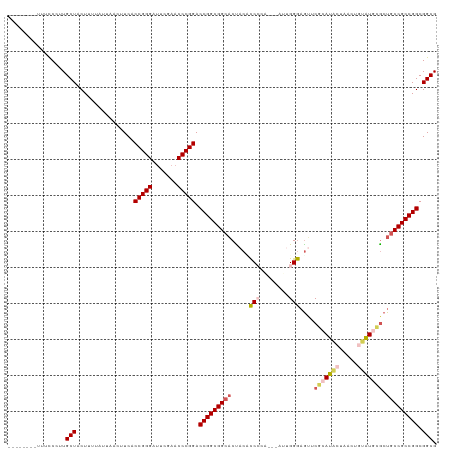
| Location | 5,726,135 – 5,726,243 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.00 |
| Shannon entropy | 0.43526 |
| G+C content | 0.59720 |
| Mean single sequence MFE | -48.00 |
| Consensus MFE | -39.52 |
| Energy contribution | -39.44 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.51 |
| SVM RNA-class probability | 0.998826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5726135 108 - 22422827 CGCCGCCGGCGGCAGCGCAUACAGUUAGUAUGCGAACUUCCAAU---CGGGAGGUAGUGCCGCCGGUCCGGGUUCGAAUCCCGGUGAAGUUAUCAAAAGUAGCAAUGGAAA-------- ((((((((((((((.(((((((.....))))))).((((((...---..))))))..))))))))))..(((.......)))))))..(((((.....)))))........-------- ( -50.70, z-score = -4.95, R) >droSim1.chrX 4465015 109 - 17042790 CGCCGCCGGCGGCAGCGCAUACAGCUAGUAUGCGAACUCCCGAU--CUCGGGUGUGGUGCCGCCGGUCCGGGUUCGAAUCCCGGUGAAGUUAAUAAAAGUAGCAACGGAAA-------- ..((((((((((((.(((((((.....))))))..((.((((..--..)))).))).))))))))))(((((.......)))))....((((.......))))...))...-------- ( -51.50, z-score = -3.70, R) >droSec1.super_4 5139713 108 + 6179234 CGCCGCCGGCGGCAGCGCAUACAGCUAGUAUGCGAACUCCCGAU---UCGGGUGUGGUGCCGCCGGUCCGGGUUCGAAUCCCGGUGAAGUUAAUAAAAGUAGCAACGGAAA-------- ..((((((((((((.(((((((.....))))))..((.((((..---.)))).))).))))))))))(((((.......)))))....((((.......))))...))...-------- ( -49.30, z-score = -3.18, R) >droYak2.chrX 19025693 113 + 21770863 CGCCGCCGGCGGCAGCACAGUUGGUUGGACUCCGAGCUCCCAAACGGAGCGGAGUAGUGCCGCCGGUCCGGGUUCGAAUCCCGGUGAAG-----UAAAGUAGCAUGGAA-AUGGAUACA ((((((((((((((...(((....))).(((((..(((((.....))))))))))..))))))))))..(((.......)))))))...-----....(((.(((....-)))..))). ( -49.70, z-score = -2.61, R) >droEre2.scaffold_4690 3074836 111 - 18748788 CGCCGCCGGCGGUAGUCCAGGUAGUUAGGCUCCGACUGCCCACA-----CGGAGUAGAGCCGCCGGUCCGGGUUCGAGUCCCGGUGAAA---GUAAAAGUAGCAUGGAACACAGACACG ((((((((((((((.(((.(((((((.(....))))))))....-----.))).)...)))))))))..(((.......)))))))...---......((.(..((.....))..))). ( -38.80, z-score = -0.53, R) >consensus CGCCGCCGGCGGCAGCGCAUACAGUUAGUAUGCGAACUCCCAAU___UCGGGAGUAGUGCCGCCGGUCCGGGUUCGAAUCCCGGUGAAGUUAAUAAAAGUAGCAACGGAAA________ .((.((((((((((.(((((((.....))))))).((.(((........))).))..))))))))))(((((.......))))).................))................ (-39.52 = -39.44 + -0.08)

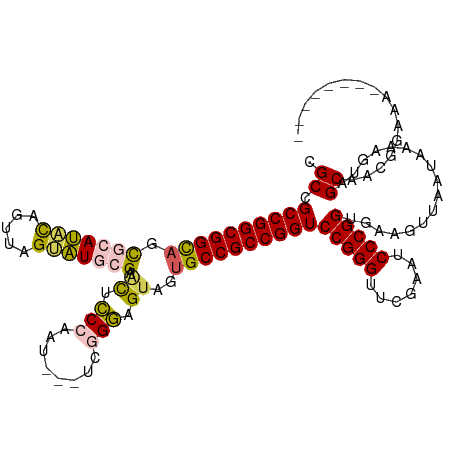
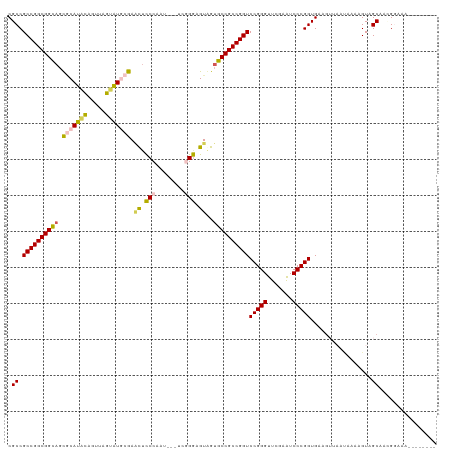
| Location | 5,726,167 – 5,726,283 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.55 |
| Shannon entropy | 0.38208 |
| G+C content | 0.62531 |
| Mean single sequence MFE | -45.51 |
| Consensus MFE | -31.68 |
| Energy contribution | -32.68 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.841055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5726167 116 + 22422827 AUUCGAACCCGGACCGGCGGCACUACCUCCCG---AUUGGAAGUUCGCAUACUAACUGUAUGCGCUGCCGCCGGCGGCGAGGGGUCAGCUCCCAGGGGAUAAACUAGAACCACACCUGC ..(((...(((..(((((((((((.((.....---...)).))).(((((((.....)))))))..))))))))))))))((((....))))((((..................)))). ( -43.77, z-score = -1.22, R) >droSim1.chrX 4465047 117 + 17042790 AUUCGAACCCGGACCGGCGGCACCACACCCGAG--AUCGGGAGUUCGCAUACUAGCUGUAUGCGCUGCCGCCGGCGGCGAGGGGACUCGCUCCAAAGGAUAAACUAGAACCUCACCUCC .(((......((((((((((((..((.((((..--..)))).)).(((((((.....))))))).)))))))))..(((((....))))))))..((......)).))).......... ( -51.60, z-score = -3.49, R) >droSec1.super_4 5139745 116 - 6179234 AUUCGAACCCGGACCGGCGGCACCACACCCGA---AUCGGGAGUUCGCAUACUAGCUGUAUGCGCUGCCGCCGGCGGCGAGGGGACUCGCUCCAAAGGAUAAACUAGAACCUCACCUCC .(((......((((((((((((..((.((((.---..)))).)).(((((((.....))))))).)))))))))..(((((....))))))))..((......)).))).......... ( -49.40, z-score = -3.12, R) >droYak2.chrX 19025727 119 - 21770863 AUUCGAACCCGGACCGGCGGCACUACUCCGCUCCGUUUGGGAGCUCGGAGUCCAACCAACUGUGCUGCCGCCGGCGGCGAGGAGACAGCGUCCAGGAGCUCAACUAGAACCUCACCUGC ........((((..(((((((((.((((((((((.....)))))..)))))..........))))))))))))).((.((((((..(((........)))...))....)))).))... ( -46.10, z-score = -0.56, R) >droEre2.scaffold_4690 3074873 114 + 18748788 ACUCGAACCCGGACCGGCGGCUCUACUCCG-----UGUGGGCAGUCGGAGCCUAACUACCUGGACUACCGCCGGCGGCGAGGUGACAGCGUCCAACUGAUAAACUAGAGCCUCACCUAC ...........(.((((((((((((((((.-----....)).))).)))))).........((....))))))))((.(((((...((.(((.....)))...))...))))).))... ( -36.70, z-score = 0.58, R) >consensus AUUCGAACCCGGACCGGCGGCACUACACCCCA___AUUGGGAGUUCGCAUACUAACUGUAUGCGCUGCCGCCGGCGGCGAGGGGACAGCCUCCAAAGGAUAAACUAGAACCUCACCUCC .(((...((((..(((((((((.....(((........)))....(((((((.....))))))).)))))))))))).).((((.....)))).............))).......... (-31.68 = -32.68 + 1.00)

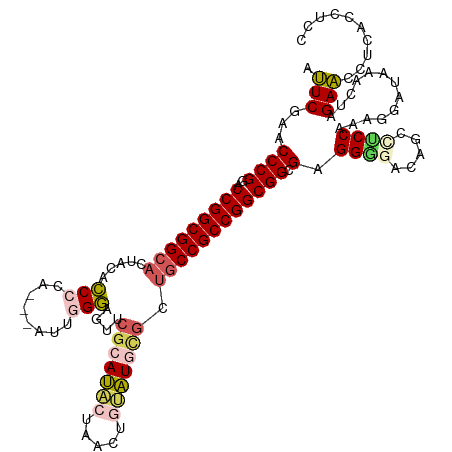
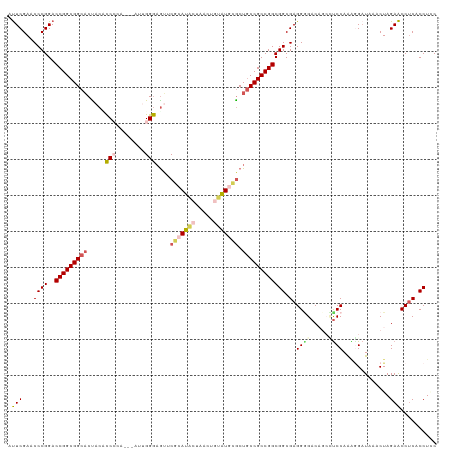
| Location | 5,726,167 – 5,726,283 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.55 |
| Shannon entropy | 0.38208 |
| G+C content | 0.62531 |
| Mean single sequence MFE | -52.44 |
| Consensus MFE | -42.25 |
| Energy contribution | -42.29 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.983237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5726167 116 - 22422827 GCAGGUGUGGUUCUAGUUUAUCCCCUGGGAGCUGACCCCUCGCCGCCGGCGGCAGCGCAUACAGUUAGUAUGCGAACUUCCAAU---CGGGAGGUAGUGCCGCCGGUCCGGGUUCGAAU ...((((.((...((((...(((....)))))))...)).))))((((((((((.(((((((.....))))))).((((((...---..))))))..))))))))))............ ( -52.90, z-score = -2.67, R) >droSim1.chrX 4465047 117 - 17042790 GGAGGUGAGGUUCUAGUUUAUCCUUUGGAGCGAGUCCCCUCGCCGCCGGCGGCAGCGCAUACAGCUAGUAUGCGAACUCCCGAU--CUCGGGUGUGGUGCCGCCGGUCCGGGUUCGAAU ((((((((((..((.((((........)))).))...))))))).(((((((((.(((((((.....))))))..((.((((..--..)))).))).)))))))))))).......... ( -58.00, z-score = -3.44, R) >droSec1.super_4 5139745 116 + 6179234 GGAGGUGAGGUUCUAGUUUAUCCUUUGGAGCGAGUCCCCUCGCCGCCGGCGGCAGCGCAUACAGCUAGUAUGCGAACUCCCGAU---UCGGGUGUGGUGCCGCCGGUCCGGGUUCGAAU ((((((((((..((.((((........)))).))...))))))).(((((((((.(((((((.....))))))..((.((((..---.)))).))).)))))))))))).......... ( -55.80, z-score = -3.03, R) >droYak2.chrX 19025727 119 + 21770863 GCAGGUGAGGUUCUAGUUGAGCUCCUGGACGCUGUCUCCUCGCCGCCGGCGGCAGCACAGUUGGUUGGACUCCGAGCUCCCAAACGGAGCGGAGUAGUGCCGCCGGUCCGGGUUCGAAU ...(((((((..(..(((.((...)).)))...)...)))))))((((((((((...(((....))).(((((..(((((.....))))))))))..))))))))))............ ( -51.30, z-score = -0.54, R) >droEre2.scaffold_4690 3074873 114 - 18748788 GUAGGUGAGGCUCUAGUUUAUCAGUUGGACGCUGUCACCUCGCCGCCGGCGGUAGUCCAGGUAGUUAGGCUCCGACUGCCCACA-----CGGAGUAGAGCCGCCGGUCCGGGUUCGAGU (.((((((((((((((........))))).))).)))))))(((((((((((((.(((.(((((((.(....))))))))....-----.))).)...)))))))))...)))...... ( -44.20, z-score = -0.38, R) >consensus GCAGGUGAGGUUCUAGUUUAUCCCUUGGACGCUGUCCCCUCGCCGCCGGCGGCAGCGCAUACAGUUAGUAUGCGAACUCCCAAU___UCGGGAGUAGUGCCGCCGGUCCGGGUUCGAAU ...(((((((((((((........)))))).......)))))))((((((((((.(((((((.....))))))).((.(((........))).))..))))))))))............ (-42.25 = -42.29 + 0.04)

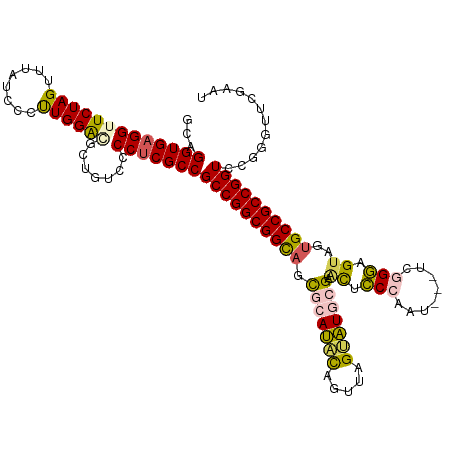
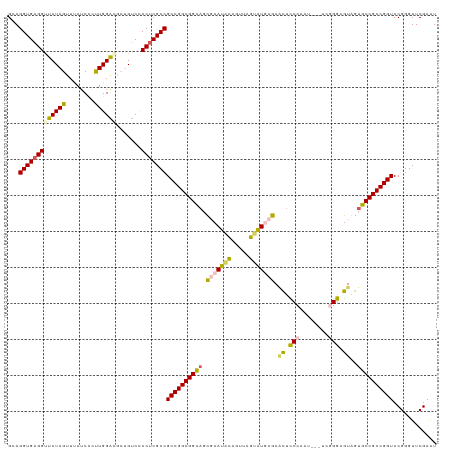
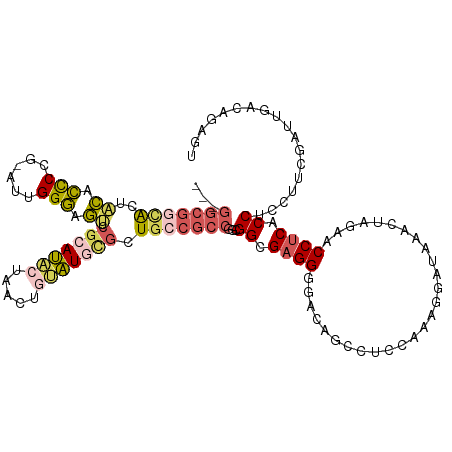
| Location | 5,726,182 – 5,726,298 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.11 |
| Shannon entropy | 0.49175 |
| G+C content | 0.59738 |
| Mean single sequence MFE | -42.53 |
| Consensus MFE | -21.95 |
| Energy contribution | -22.91 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.587794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5726182 116 + 22422827 ---GGCGGCACUACCUCCCG-AUUGGAAGUUCGCAUACUAACUGUAUGCGCUGCCGCCGGCGGCGAGGGGUCAGCUCCCAGGGGAUAAACUAGAACCACACCUGCUUCGAUUGACAGAUU ---(((((((((.((.....-...)).))).(((((((.....)))))))..))))))(((((....((.((((.(((....)))....)).)).))....))))).............. ( -38.50, z-score = -0.43, R) >droSim1.chrX 4465062 117 + 17042790 ---GGCGGCACCACACCCGAGAUCGGGAGUUCGCAUACUAGCUGUAUGCGCUGCCGCCGGCGGCGAGGGGACUCGCUCCAAAGGAUAAACUAGAACCUCACCUCCUUCGAUUGACAGAGU ---(((((((..((.((((....)))).)).(((((((.....))))))).)))))))((.((((((....)))))))).(((((.................)))))............. ( -48.73, z-score = -2.97, R) >droSec1.super_4 5139760 116 - 6179234 ---GGCGGCACCACACCCGA-AUCGGGAGUUCGCAUACUAGCUGUAUGCGCUGCCGCCGGCGGCGAGGGGACUCGCUCCAAAGGAUAAACUAGAACCUCACCUCCUUCGAUUGACAGAGU ---(((((((..((.((((.-..)))).)).(((((((.....))))))).)))))))((.((((((....)))))))).(((((.................)))))............. ( -46.53, z-score = -2.58, R) >droYak2.chrX 19025742 119 - 21770863 GGCGGCACUACUCCGCUCCG-UUUGGGAGCUCGGAGUCCAACCAACUGUGCUGCCGCCGGCGGCGAGGAGACAGCGUCCAGGAGCUCAACUAGAACCUCACCUGCUUCGAUUGCCACAAA ((((((((.((((((((((.-....)))))..)))))..........))))))))...((((((((((...(((.((..(((..((.....))..))).)))))))))).)))))..... ( -45.70, z-score = -1.01, R) >droEre2.scaffold_4690 3074888 106 + 18748788 -----GGCGGCUCUACUCCG-UGUGGGCAGUCGGAGCCUAACUACCUGGACUACCGCCGGCGGCGAGGUGACAGCGUCCAACUGAUAAACUAGAGCCUCACCUACUUCGAUU-------- -----((((((((((((((.-....)).))).))))))........(((((...((((...)))).(....)...)))))..............)))...............-------- ( -33.20, z-score = 0.45, R) >consensus ___GGCGGCACUACACCCCG_AUUGGGAGUUCGCAUACUAACUGUAUGCGCUGCCGCCGGCGGCGAGGGGACAGCCUCCAAAGGAUAAACUAGAACCUCACCUCCUUCGAUUGACAGAGU ...(((((((..((.(((......))).)).(((((((.....))))))).)))))))...((.((((...........................)))).)).................. (-21.95 = -22.91 + 0.96)

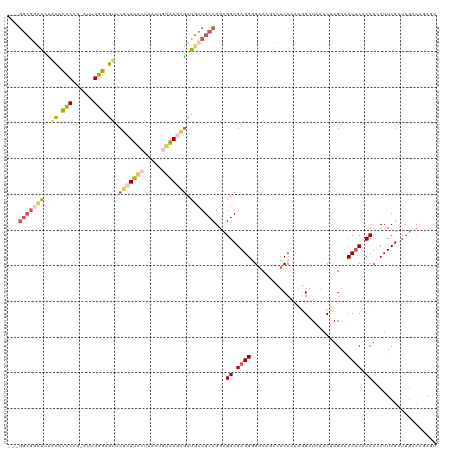
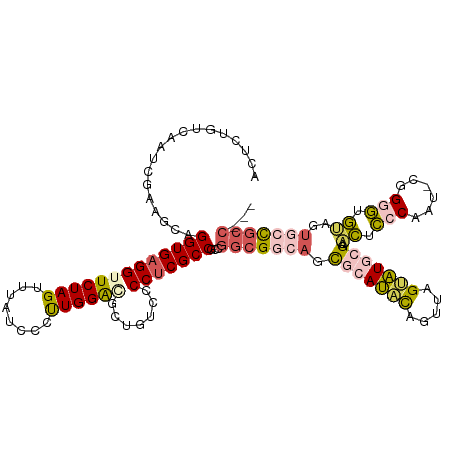
| Location | 5,726,182 – 5,726,298 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.11 |
| Shannon entropy | 0.49175 |
| G+C content | 0.59738 |
| Mean single sequence MFE | -46.78 |
| Consensus MFE | -28.23 |
| Energy contribution | -29.39 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.960867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5726182 116 - 22422827 AAUCUGUCAAUCGAAGCAGGUGUGGUUCUAGUUUAUCCCCUGGGAGCUGACCCCUCGCCGCCGGCGGCAGCGCAUACAGUUAGUAUGCGAACUUCCAAU-CGGGAGGUAGUGCCGCC--- .((((((........))))))(((((....(((..(((....)))...))).....))))).(((((((.(((((((.....))))))).((((((...-..))))))..)))))))--- ( -46.90, z-score = -2.48, R) >droSim1.chrX 4465062 117 - 17042790 ACUCUGUCAAUCGAAGGAGGUGAGGUUCUAGUUUAUCCUUUGGAGCGAGUCCCCUCGCCGCCGGCGGCAGCGCAUACAGCUAGUAUGCGAACUCCCGAUCUCGGGUGUGGUGCCGCC--- ...............((.(((((((..((.((((........)))).))...))))))).))(((((((.(((((((.....))))))..((.((((....)))).))).)))))))--- ( -51.90, z-score = -2.93, R) >droSec1.super_4 5139760 116 + 6179234 ACUCUGUCAAUCGAAGGAGGUGAGGUUCUAGUUUAUCCUUUGGAGCGAGUCCCCUCGCCGCCGGCGGCAGCGCAUACAGCUAGUAUGCGAACUCCCGAU-UCGGGUGUGGUGCCGCC--- ...............((.(((((((..((.((((........)))).))...))))))).))(((((((.(((((((.....))))))..((.((((..-.)))).))).)))))))--- ( -49.70, z-score = -2.49, R) >droYak2.chrX 19025742 119 + 21770863 UUUGUGGCAAUCGAAGCAGGUGAGGUUCUAGUUGAGCUCCUGGACGCUGUCUCCUCGCCGCCGGCGGCAGCACAGUUGGUUGGACUCCGAGCUCCCAAA-CGGAGCGGAGUAGUGCCGCC ...(((((((((((..((((...(((((.....)))))))))...((((((.((........)).))))))....))))))..(((((..(((((....-.))))))))))...))))). ( -47.70, z-score = -0.37, R) >droEre2.scaffold_4690 3074888 106 - 18748788 --------AAUCGAAGUAGGUGAGGCUCUAGUUUAUCAGUUGGACGCUGUCACCUCGCCGCCGGCGGUAGUCCAGGUAGUUAGGCUCCGACUGCCCACA-CGGAGUAGAGCCGCC----- --------..........(((..(((((((.((..(((((((((.(((..(((((.((((....)))).....)))).)...))))))))))).....)-..)).))))))))))----- ( -37.70, z-score = -0.59, R) >consensus ACUCUGUCAAUCGAAGCAGGUGAGGUUCUAGUUUAUCCCUUGGACGCUGUCCCCUCGCCGCCGGCGGCAGCGCAUACAGUUAGUAUGCGAACUCCCAAU_CGGGGUGUAGUGCCGCC___ ..................(((((((((((((........)))))).......)))))))...(((((((.(((((((.....))))))).((.(((......))).))..)))))))... (-28.23 = -29.39 + 1.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:18:51 2011