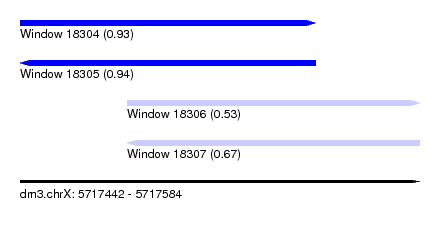
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,717,442 – 5,717,584 |
| Length | 142 |
| Max. P | 0.936604 |

| Location | 5,717,442 – 5,717,547 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 70.80 |
| Shannon entropy | 0.52928 |
| G+C content | 0.42736 |
| Mean single sequence MFE | -28.55 |
| Consensus MFE | -16.67 |
| Energy contribution | -16.37 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.930706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
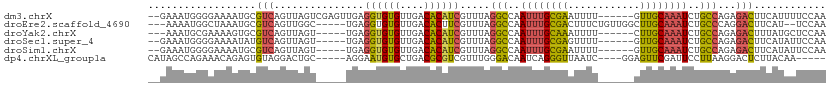
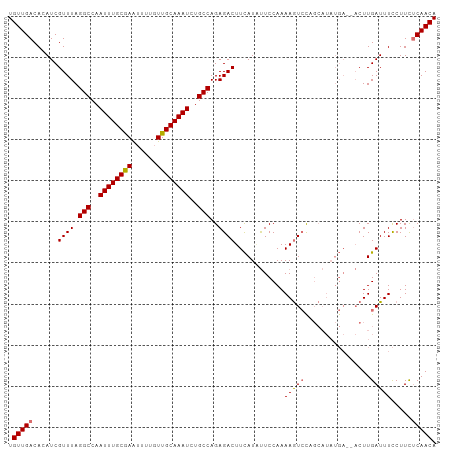
>dm3.chrX 5717442 105 + 22422827 --GAAAUGGGGAAAAUGCGUCAGUUAGUCGAGUUGAGGUGUGUUGACACAUCGUUUAGGCCAAUUUGCGAAUUUU------GUUGCAAAUCUGCCAGAGACUUCAUUUUCCAA --.......((((((((.(((...............((((((....)))))).....(((..((((((((.....------.))))))))..)))...)))..)))))))).. ( -33.50, z-score = -2.70, R) >droEre2.scaffold_4690 3066281 103 + 18748788 ---AAAAUGGCUAAAUGCGUCAGUUGGC-----UGAGGUGUGUUGACACUUCGUUUAGGCCAAUUUGCGACUUUCUGUUGGCUUGCAAAUCUGCCCAGGACUUCAU--UCCAA ---....(((...(((((((.(((((((-----((((((((....))))))).....)))))))).)))...(((((..(((..........))))))))...)))--)))). ( -29.40, z-score = -0.83, R) >droYak2.chrX 19015697 99 - 21770863 ---AAAUGCGAAAAGUGCGUCAGUUAGU-----UGAGGUGUGUUGACACAUCGUUUAGGCCAAUUUGCAAAUUUU------CUUGCAAAUCUGCCAGAGACUUUAUGCUCCAA ---....(((.(((((.(.((.......-----...((((((....)))))).....(((..((((((((.....------.))))))))..))).)))))))).)))..... ( -28.00, z-score = -1.88, R) >droSec1.super_4 5131171 100 - 6179234 --GAAAUGGGGAAAAUAUGUCAGUUAGU-----UGAGGUGUGUUGACACAUCGUUUAGGCCAAUUUGCGAGUUUU------GUUGCAAAUCUGCCAGAGACUUCAUAUUCCAA --.......(((..(((((..((((...-----...((((((....)))))).....(((..((((((((.....------.))))))))..)))...)))).)))))))).. ( -28.60, z-score = -2.10, R) >droSim1.chrX 4456271 100 + 17042790 --GAAAUGGGGAAAAUGCGUCAGUUAGU-----UGAGGUGUGUUGACACAUCGUUUAGGCCAAUUUGCGAAUUUU------GUUGCAAAUCUGCCAGAGACUUCAUAUUCCAA --.......((((.(((.(((.......-----...((((((....)))))).....(((..((((((((.....------.))))))))..)))...)))..))).)))).. ( -30.00, z-score = -2.24, R) >dp4.chrXL_group1a 7059313 99 - 9151740 CAUAGCCAGAAACAGAGUGUAGGACUGC-----AGGAAUGUGCUGACGCGUCGUUUGGGACAAUCAGGGUUAAUC----GGAGUUCGAUUCCUUAAGGACUCUUACAA----- ..(((((.........((((((.((...-----......)).)).))))(((......)))......)))))...----(((((((..........))))))).....----- ( -21.80, z-score = 0.91, R) >consensus __GAAAUGGGGAAAAUGCGUCAGUUAGU_____UGAGGUGUGUUGACACAUCGUUUAGGCCAAUUUGCGAAUUUU______GUUGCAAAUCUGCCAGAGACUUCAUAUUCCAA ..................(((...............((((((....)))))).....(((..((((((((............))))))))..)))...)))............ (-16.67 = -16.37 + -0.30)
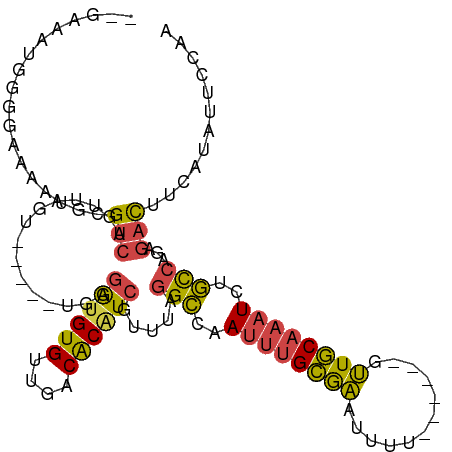
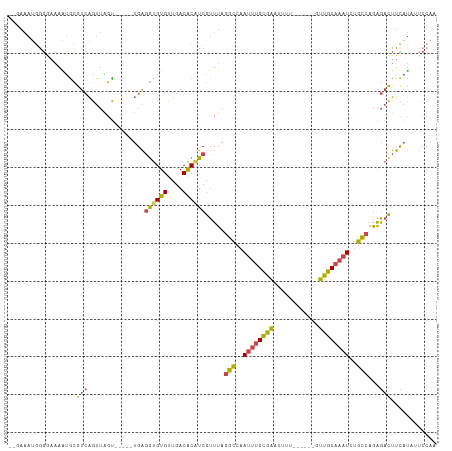
| Location | 5,717,442 – 5,717,547 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 70.80 |
| Shannon entropy | 0.52928 |
| G+C content | 0.42736 |
| Mean single sequence MFE | -23.31 |
| Consensus MFE | -11.79 |
| Energy contribution | -13.12 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.936604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5717442 105 - 22422827 UUGGAAAAUGAAGUCUCUGGCAGAUUUGCAAC------AAAAUUCGCAAAUUGGCCUAAACGAUGUGUCAACACACCUCAACUCGACUAACUGACGCAUUUUCCCCAUUUC-- ..((((((((..(((...(((.(((((((...------.......))))))).))).....((.((((....)))).)).............))).)))))))).......-- ( -27.70, z-score = -3.47, R) >droEre2.scaffold_4690 3066281 103 - 18748788 UUGGA--AUGAAGUCCUGGGCAGAUUUGCAAGCCAACAGAAAGUCGCAAAUUGGCCUAAACGAAGUGUCAACACACCUCA-----GCCAACUGACGCAUUUAGCCAUUUU--- ..(((--(((..(((.(((((.(((((((................))))))).)))))...((.((((....)))).)).-----.......)))((.....))))))))--- ( -23.09, z-score = -0.06, R) >droYak2.chrX 19015697 99 + 21770863 UUGGAGCAUAAAGUCUCUGGCAGAUUUGCAAG------AAAAUUUGCAAAUUGGCCUAAACGAUGUGUCAACACACCUCA-----ACUAACUGACGCACUUUUCGCAUUU--- .....((..(((((....(((.((((((((((------....)))))))))).)))........((((((..........-----......)))))))))))..))....--- ( -25.29, z-score = -2.17, R) >droSec1.super_4 5131171 100 + 6179234 UUGGAAUAUGAAGUCUCUGGCAGAUUUGCAAC------AAAACUCGCAAAUUGGCCUAAACGAUGUGUCAACACACCUCA-----ACUAACUGACAUAUUUUCCCCAUUUC-- ..((((((((.(((....(((.(((((((...------.......))))))).))).....((.((((....)))).)).-----....)))..)))))..))).......-- ( -21.90, z-score = -2.35, R) >droSim1.chrX 4456271 100 - 17042790 UUGGAAUAUGAAGUCUCUGGCAGAUUUGCAAC------AAAAUUCGCAAAUUGGCCUAAACGAUGUGUCAACACACCUCA-----ACUAACUGACGCAUUUUCCCCAUUUC-- ..((((.(((..(((...(((.(((((((...------.......))))))).))).....((.((((....)))).)).-----.......))).))).)))).......-- ( -24.40, z-score = -2.72, R) >dp4.chrXL_group1a 7059313 99 + 9151740 -----UUGUAAGAGUCCUUAAGGAAUCGAACUCC----GAUUAACCCUGAUUGUCCCAAACGACGCGUCAGCACAUUCCU-----GCAGUCCUACACUCUGUUUCUGGCUAUG -----.......((((....((((((((.....)----))......(((((((((......)))).)))))....)))))-----((((.........))))....))))... ( -17.50, z-score = 0.14, R) >consensus UUGGAAUAUGAAGUCUCUGGCAGAUUUGCAAC______AAAAUUCGCAAAUUGGCCUAAACGAUGUGUCAACACACCUCA_____ACUAACUGACGCAUUUUCCCCAUUUC__ ..................(((.(((((((................))))))).))).....(((((((((.....................)))))))))............. (-11.79 = -13.12 + 1.34)
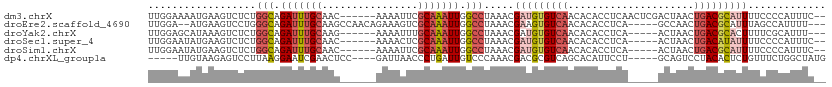
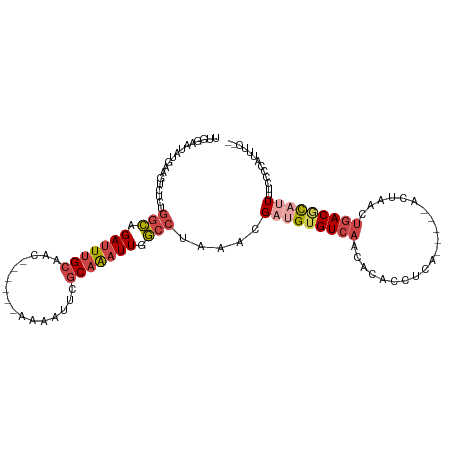
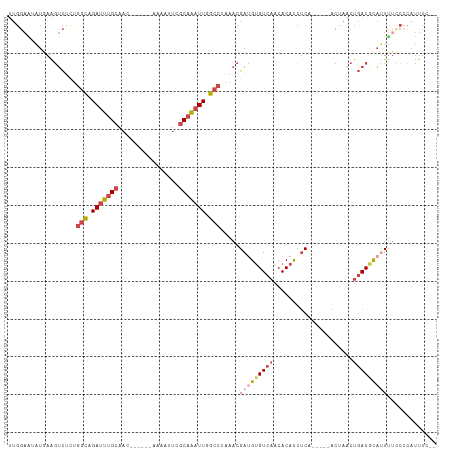
| Location | 5,717,480 – 5,717,584 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 87.54 |
| Shannon entropy | 0.20228 |
| G+C content | 0.37736 |
| Mean single sequence MFE | -22.28 |
| Consensus MFE | -16.99 |
| Energy contribution | -17.55 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.531437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
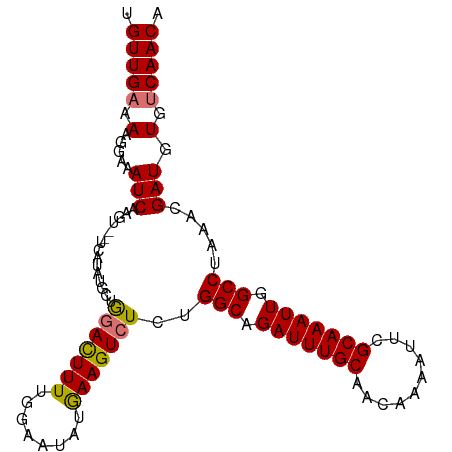
>dm3.chrX 5717480 104 + 22422827 UGUUGACACAUCGUUUAGGCCAAUUUGCGAAUUUUGUUGCAAAUCUGCCAGAGACUUCAUUUUCCAAAAGUCUUGCAUAUGAGAACUUGAUUUCCUUUUCAACA .(((((....((((...(((..((((((((......))))))))..))).(((((((..........)))))))....))))(((......)))....))))). ( -23.20, z-score = -1.57, R) >droYak2.chrX 19015729 92 - 21770863 UGUUGACACAUCGUUUAGGCCAAUUUGCAAAUUUUCUUGCAAAUCUGCCAGAGACUUUAUGCUCCAAAAAUGC------------CUUGAUUUUCUUUGCAACA .((((.((.........(((..((((((((......))))))))..)))(((((.((...((.........))------------...)).))))).)))))). ( -20.60, z-score = -1.78, R) >droSec1.super_4 5131204 102 - 6179234 UGUUGACACAUCGUUUAGGCCAAUUUGCGAGUUUUGUUGCAAAUCUGCCAGAGACUUCAUAUUCCAAAAGUCCAGCAUAUGA--ACUUGAUUUCCUUCUCAACA .(((((......((((((((..((((((((......))))))))..))).(.(((((..........))))))......)))--))..((......))))))). ( -22.60, z-score = -1.56, R) >droSim1.chrX 4456304 102 + 17042790 UGUUGACACAUCGUUUAGGCCAAUUUGCGAAUUUUGUUGCAAAUCUGCCAGAGACUUCAUAUUCCAAAAGUCCAGCAUAUGA--ACUUGAUUUCCUUCUCAACA .(((((......((((((((..((((((((......))))))))..))).(.(((((..........))))))......)))--))..((......))))))). ( -22.70, z-score = -1.93, R) >consensus UGUUGACACAUCGUUUAGGCCAAUUUGCGAAUUUUGUUGCAAAUCUGCCAGAGACUUCAUAUUCCAAAAGUCCAGCAUAUGA__ACUUGAUUUCCUUCUCAACA .(((((...........(((..((((((((......))))))))..)))...(((((..........)))))..........................))))). (-16.99 = -17.55 + 0.56)

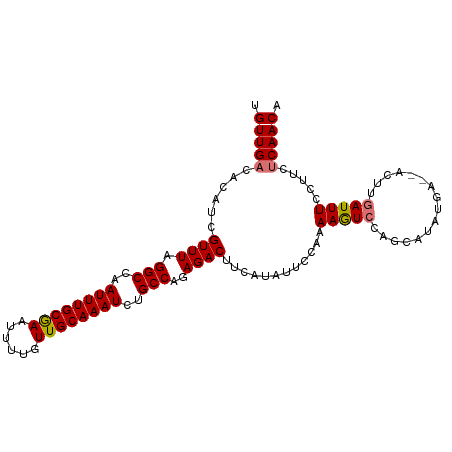
| Location | 5,717,480 – 5,717,584 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 87.54 |
| Shannon entropy | 0.20228 |
| G+C content | 0.37736 |
| Mean single sequence MFE | -24.73 |
| Consensus MFE | -21.56 |
| Energy contribution | -21.50 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.668355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
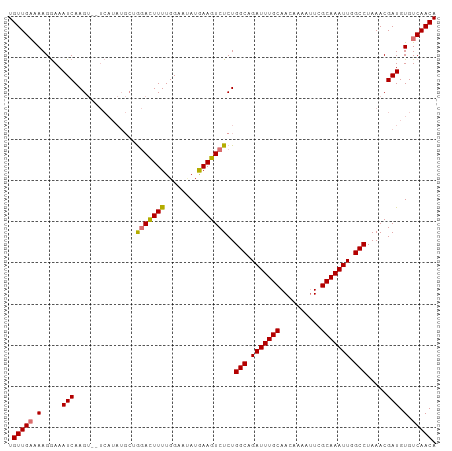
>dm3.chrX 5717480 104 - 22422827 UGUUGAAAAGGAAAUCAAGUUCUCAUAUGCAAGACUUUUGGAAAAUGAAGUCUCUGGCAGAUUUGCAACAAAAUUCGCAAAUUGGCCUAAACGAUGUGUCAACA .(((((...((((......))))(((((.(.(((((((........)))))))..(((.(((((((..........))))))).))).....))))))))))). ( -25.30, z-score = -1.63, R) >droYak2.chrX 19015729 92 + 21770863 UGUUGCAAAGAAAAUCAAG------------GCAUUUUUGGAGCAUAAAGUCUCUGGCAGAUUUGCAAGAAAAUUUGCAAAUUGGCCUAAACGAUGUGUCAACA .((((((......(((...------------........(((((.....).))))(((.((((((((((....)))))))))).))).....))).)).)))). ( -22.40, z-score = -1.01, R) >droSec1.super_4 5131204 102 + 6179234 UGUUGAGAAGGAAAUCAAGU--UCAUAUGCUGGACUUUUGGAAUAUGAAGUCUCUGGCAGAUUUGCAACAAAACUCGCAAAUUGGCCUAAACGAUGUGUCAACA .(((((.(.....(((...(--((((((.(..(....)..).)))))))((....(((.(((((((..........))))))).)))...))))).).))))). ( -25.60, z-score = -1.39, R) >droSim1.chrX 4456304 102 - 17042790 UGUUGAGAAGGAAAUCAAGU--UCAUAUGCUGGACUUUUGGAAUAUGAAGUCUCUGGCAGAUUUGCAACAAAAUUCGCAAAUUGGCCUAAACGAUGUGUCAACA .(((((.(.....(((...(--((((((.(..(....)..).)))))))((....(((.(((((((..........))))))).)))...))))).).))))). ( -25.60, z-score = -1.40, R) >consensus UGUUGAAAAGGAAAUCAAGU__UCAUAUGCUGGACUUUUGGAAUAUGAAGUCUCUGGCAGAUUUGCAACAAAAUUCGCAAAUUGGCCUAAACGAUGUGUCAACA .(((((.(.....(((...............(((((((........)))))))..(((.(((((((..........))))))).))).....))).).))))). (-21.56 = -21.50 + -0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:18:43 2011