| Sequence ID | dm3.chr2L |
|---|---|
| Location | 901,461 – 901,549 |
| Length | 88 |
| Max. P | 0.967660 |

| Location | 901,461 – 901,549 |
|---|---|
| Length | 88 |
| Sequences | 10 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 82.78 |
| Shannon entropy | 0.38712 |
| G+C content | 0.43678 |
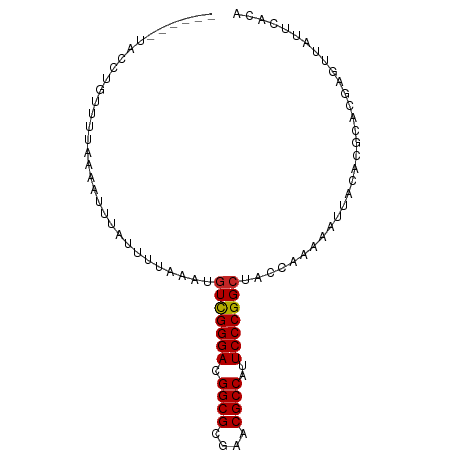
| Mean single sequence MFE | -20.23 |
| Consensus MFE | -17.57 |
| Energy contribution | -17.58 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.62 |
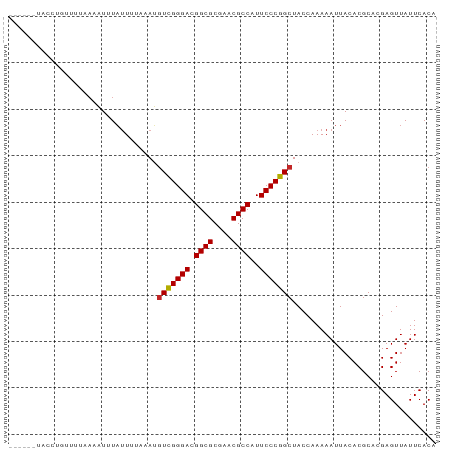
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.967660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
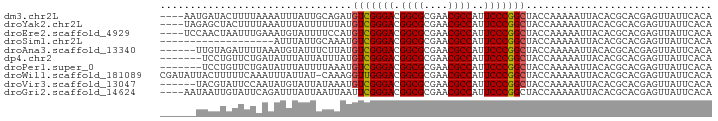
>dm3.chr2L 901461 88 + 23011544 ----AAUGAUACUUUUAAAAUUUAUUGCAGAUGUCGGGACGGCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACA ----..(((.((((.........(((......(((((((.((((....))))..))))))).......))).........))))...))).. ( -20.69, z-score = -1.63, R) >droYak2.chr2L 887356 88 + 22324452 ----UAGAGCUACUUUUAAAUUUAUUUUUUAUGUCGGGACGGCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACA ----..(((..((((........((((((...(((((((.((((....))))..)))))))....)))))).........))))..)))... ( -23.53, z-score = -3.23, R) >droEre2.scaffold_4929 963507 88 + 26641161 ----UCCAACUAAUUUGAAAUGUAUUUUCCAUGUCGGGACGGCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACA ----......(((((((...((((((((....(((((((.((((....))))..))))))).....)))).))))....)))))))...... ( -23.40, z-score = -2.50, R) >droSim1.chr2L 890061 73 + 22036055 -------------------AUUUAUUGCAAAUGUCGGGACGGCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACA -------------------.............(((((((.((((....))))..)))))))............................... ( -18.20, z-score = -1.20, R) >droAna3.scaffold_13340 22991005 86 + 23697760 ------UUGUAGAUUUUAAAUGUAUUUCUUAUGUCGGGACGGCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACA ------.(((((.((((....(((.....)))(((((((.((((....))))..)))))))....)))).)))))................. ( -20.10, z-score = -1.18, R) >dp4.chr2 12924947 85 + 30794189 -------UCCUGUUCUGAUAUUUAUUAUUUAUGUCGGGACGGCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACA -------((.(((...((((.....))))...(((((((.((((....))))..)))))))...............))).)).......... ( -18.70, z-score = -0.88, R) >droPer1.super_0 4291580 85 - 11822988 -------UCCUGUUCUGAUAUUUAUUUUAAAUGUCGGGACGGCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACA -------....(((((((((((((...)))))))))))))((((....)))).....((((...............)).))........... ( -20.76, z-score = -1.59, R) >droWil1.scaffold_181089 5881044 91 - 12369635 CGAUAUUACUUUUUCAAAUUUAUUAU-CAAAGGUUGGGACGGCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACA .((((.((..((....))..)).)))-)...((((((((.((((....))))..)))))))).............................. ( -18.70, z-score = -1.45, R) >droVir3.scaffold_13047 18360234 86 + 19223366 ------UACGUAUUCCAAUAUGUAUUAUAAAUGUCGGGACGGCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACA ------(((((((....)))))))........(((((((.((((....))))..)))))))............................... ( -21.70, z-score = -1.72, R) >droGri2.scaffold_14624 1609949 88 + 4233967 ----AAUAAUUGUAUUCAGAUUUAUUAAUUAAUUCGGGACGGCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACA ----.........................((((((((((.((((....))))..))))((...))...............))))))...... ( -16.50, z-score = -0.80, R) >consensus ______UACCUGUUUUAAAAUUUAUUUUAAAUGUCGGGACGGCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACA ................................(((((((.((((....))))..)))))))............................... (-17.57 = -17.58 + 0.01)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:07:17 2011