
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,688,478 – 5,688,590 |
| Length | 112 |
| Max. P | 0.624332 |

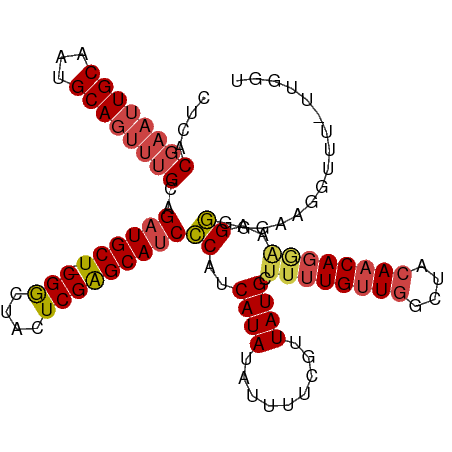
| Location | 5,688,478 – 5,688,586 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 86.20 |
| Shannon entropy | 0.26665 |
| G+C content | 0.44994 |
| Mean single sequence MFE | -32.62 |
| Consensus MFE | -19.39 |
| Energy contribution | -20.87 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.573753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
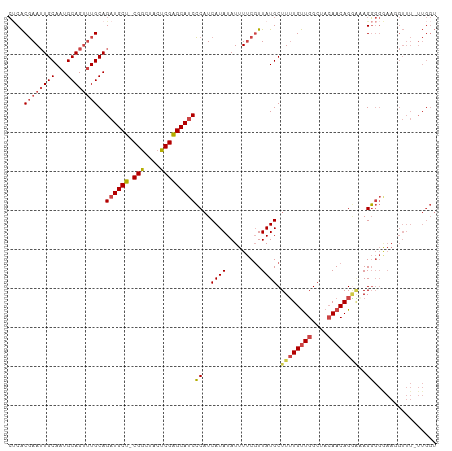
>dm3.chrX 5688478 108 - 22422827 CACACGAAUUGCAAUGCAGUUUGCAGAUGCU-CGGCUACUCGAGCAUCCCAUCAUAUAUUUUCGUUAUGCUUUUGUUGGCUACAACAGGAAAGGGGGAAGGUUU-UUGGU ....((((((((...))))))))..((((((-(((....)))))))))..((((...((((((.((....((((((((....))))))))...)).))))))..-.)))) ( -32.90, z-score = -2.23, R) >droSim1.chrX 4428925 108 - 17042790 CUCACGAAUUGCAAUGCAGUUUGCAGAUGCU-CGGCUACUCGAGCAUCCCAUCAUAUAUUUUCGUUAUGCUUUUGUUGGCUACAACAGGGUAGGGGGAAUGUUU-UUGGU ....((((((((...))))))))..((((((-(((....)))))))))(((....(((((..(.((((.((.((((.....)))).)).)))).)..)))))..-.))). ( -36.90, z-score = -3.30, R) >droSec1.super_4 5103433 108 + 6179234 CUCACGAAUUGCAAUGCAGUUUGCAGAUGCU-CGGCUACUCGAGCAUCCCAUCAUAUAUUUUCGUUAUGCUUUUGUUGGCUACAACAGGGUAGGGGGAAUGUUU-UUGGU ....((((((((...))))))))..((((((-(((....)))))))))(((....(((((..(.((((.((.((((.....)))).)).)))).)..)))))..-.))). ( -36.90, z-score = -3.30, R) >droYak2.chrX 18985621 109 + 21770863 CUCACGAAUUGCAAUGCAGUUUGCAGAUGCU-CGGCUACUCGAGCAUCCCAUCAUAUAUUUUCGUUAUGCUUUUGUUGGCUACAACAGGAAAGGGGAAAGGUUUGUUGGU ....((((((((...))))))))..((((((-(((....)))))))))(((.((.((.(((((.((....((((((((....)))))))).)).))))).)).)).))). ( -32.90, z-score = -1.89, R) >droEre2.scaffold_4690 3037146 108 - 18748788 CUCACGAAUUGCAAUGCAGUUUGCAGAUGCU-CGGCUACUCGAGCAUCCCAUCAUAUAUUUUCGUUAUGCUUUUGUUGGCUACAACAGAAAAGGGGAAAGGUUU-UUGGU ....((((((((...))))))))..((((((-(((....)))))))))(((.......(((((.((....((((((((....)))))))).)).))))).....-.))). ( -31.12, z-score = -1.85, R) >droAna3.scaffold_13117 1123104 90 - 5790199 GCCGCCAAGUGCAAUGCAG-AUGCAGAUGCUCCGACUACUCGGGCACCGC-UCAUAUAUUUCCUUUAUGCUUGUGCUGGCU--AACAGUUUAGU---------------- ...((((.(..(((.((.(-(.((.(.(((.((((....)))))))).))-))..(((.......))))))))..))))).--...........---------------- ( -25.00, z-score = -0.30, R) >consensus CUCACGAAUUGCAAUGCAGUUUGCAGAUGCU_CGGCUACUCGAGCAUCCCAUCAUAUAUUUUCGUUAUGCUUUUGUUGGCUACAACAGGAAAGGGGGAAGGUUU_UUGGU ((((((((.(((((......)))))((((((.(((....)))))))))............))))).....((((((((....))))))))..)))............... (-19.39 = -20.87 + 1.48)

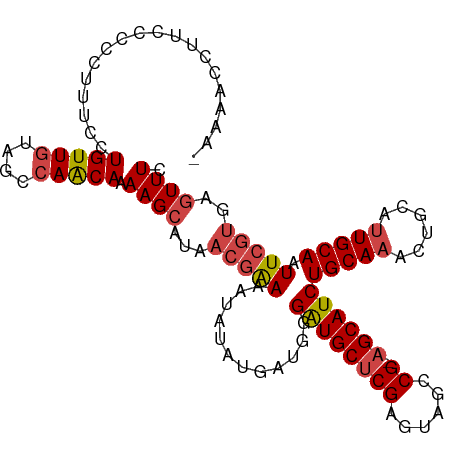
| Location | 5,688,482 – 5,688,590 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 86.13 |
| Shannon entropy | 0.27038 |
| G+C content | 0.44038 |
| Mean single sequence MFE | -27.37 |
| Consensus MFE | -18.20 |
| Energy contribution | -18.95 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.624332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
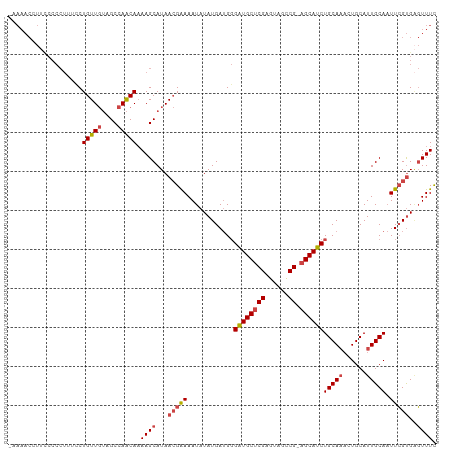
>dm3.chrX 5688482 108 + 22422827 -AAAACCUUCCCCCUUUCCUGUUGUAGCCAACAAAAGCAUAACGAAAAUAUAUGAUGGGAUGCUCGAGUAGCCG-AGCAUCUGCAAACUGCAUUGCAAUUCGUGUGUUUC -..................((((((.((........)))))))).((((((((((..(((((((((......))-)))))))((((......))))...)))))))))). ( -29.50, z-score = -2.99, R) >droSim1.chrX 4428929 108 + 17042790 -AAAACAUUCCCCCUACCCUGUUGUAGCCAACAAAAGCAUAACGAAAAUAUAUGAUGGGAUGCUCGAGUAGCCG-AGCAUCUGCAAACUGCAUUGCAAUUCGUGAGUUUC -..................(((((....))))).((((...(((((...........(((((((((......))-)))))))((((......))))..)))))..)))). ( -26.70, z-score = -2.15, R) >droSec1.super_4 5103437 108 - 6179234 -AAAACAUUCCCCCUACCCUGUUGUAGCCAACAAAAGCAUAACGAAAAUAUAUGAUGGGAUGCUCGAGUAGCCG-AGCAUCUGCAAACUGCAUUGCAAUUCGUGAGUUUC -..................(((((....))))).((((...(((((...........(((((((((......))-)))))))((((......))))..)))))..)))). ( -26.70, z-score = -2.15, R) >droYak2.chrX 18985625 109 - 21770863 ACAAACCUUUCCCCUUUCCUGUUGUAGCCAACAAAAGCAUAACGAAAAUAUAUGAUGGGAUGCUCGAGUAGCCG-AGCAUCUGCAAACUGCAUUGCAAUUCGUGAGUUUC ...................(((((....))))).((((...(((((...........(((((((((......))-)))))))((((......))))..)))))..)))). ( -26.70, z-score = -1.76, R) >droEre2.scaffold_4690 3037150 108 + 18748788 -AAAACCUUUCCCCUUUUCUGUUGUAGCCAACAAAAGCAUAACGAAAAUAUAUGAUGGGAUGCUCGAGUAGCCG-AGCAUCUGCAAACUGCAUUGCAAUUCGUGAGUUUC -.((((........(((((.(((((.((........))))))))))))..(((((..(((((((((......))-)))))))((((......))))...))))).)))). ( -27.00, z-score = -1.90, R) >droAna3.scaffold_13117 1123108 90 + 5790199 ----------------AACUGUU--AGCCAGCACAAGCAUAAAGGAAAUAUAUGA-GCGGUGCCCGAGUAGUCGGAGCAUCUGCAU-CUGCAUUGCACUUGGCGGCUUUU ----------------.......--((((.((.((((((((.........)))).-(((((((((((....)))).)))))(((..-..)))..)).))))))))))... ( -27.60, z-score = -0.56, R) >consensus _AAAACCUUCCCCCUUUCCUGUUGUAGCCAACAAAAGCAUAACGAAAAUAUAUGAUGGGAUGCUCGAGUAGCCG_AGCAUCUGCAAACUGCAUUGCAAUUCGUGAGUUUC ...................(((((....))))).((((...(((((............((((((((......)).))))))(((((......))))).)))))..)))). (-18.20 = -18.95 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:18:36 2011