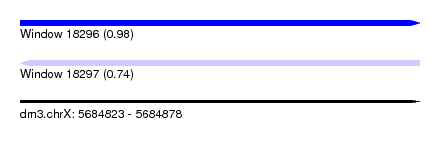
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,684,823 – 5,684,878 |
| Length | 55 |
| Max. P | 0.981775 |

| Location | 5,684,823 – 5,684,878 |
|---|---|
| Length | 55 |
| Sequences | 5 |
| Columns | 61 |
| Reading direction | forward |
| Mean pairwise identity | 75.26 |
| Shannon entropy | 0.41930 |
| G+C content | 0.48197 |
| Mean single sequence MFE | -16.34 |
| Consensus MFE | -12.57 |
| Energy contribution | -13.33 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.981775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
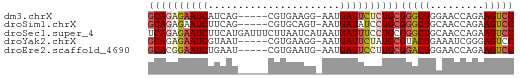
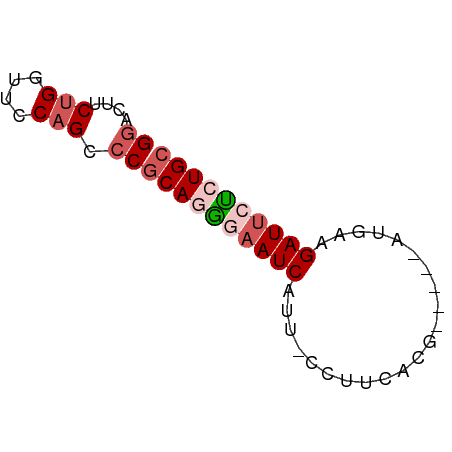
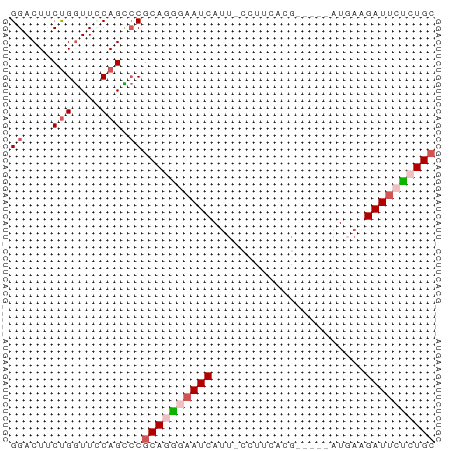
>dm3.chrX 5684823 55 + 22422827 GCAGAGAAUCAUCAG-----CGUGAAGG-AAUGAUUCUCUGCGGGCUGGAACCAGAAGUCC ((((((((((((...-----(......)-.))))))))))))(((((.........))))) ( -22.40, z-score = -3.41, R) >droSim1.chrX 4425310 55 + 17042790 GCAGAGAAUCUUCAG-----CGUGCAGU-AAUGAUAUCCUGCGGGCUGCAACCAGAAGUCC ((((.(((((....(-----(.....))-...))).))))))(((((.........))))) ( -12.90, z-score = -0.12, R) >droSec1.super_4 5099861 61 - 6179234 UCAGAGAAUCUUCAUGAUUUCUUAAUCAUAAUGAUUUCCUGCGGGCUGCAACCAGAAGUCC .(((..((((...((((((....))))))...))))..))).(((((.........))))) ( -15.60, z-score = -1.93, R) >droYak2.chrX 18981891 55 - 21770863 GCAGAGAAUCGUAAU-----CGUGAAGG-AAUGAUUCUAUGCGUACUGAAAUCGGGAGUCC (((.((((((((..(-----(......)-))))))))).)))...(((....)))...... ( -13.80, z-score = -1.48, R) >droEre2.scaffold_4690 3033434 55 + 18748788 GCACGGAAUCUGAAU-----CGUGAAUG-AAUGAUUCCUUGCGGACUGGAACCAGAAGUCC (((.((((((....(-----((....))-)..)))))).)))(((((.........))))) ( -17.00, z-score = -2.60, R) >consensus GCAGAGAAUCUUCAU_____CGUGAAGG_AAUGAUUCCCUGCGGGCUGCAACCAGAAGUCC ((((((((((......................))))))))))(((((.........))))) (-12.57 = -13.33 + 0.76)
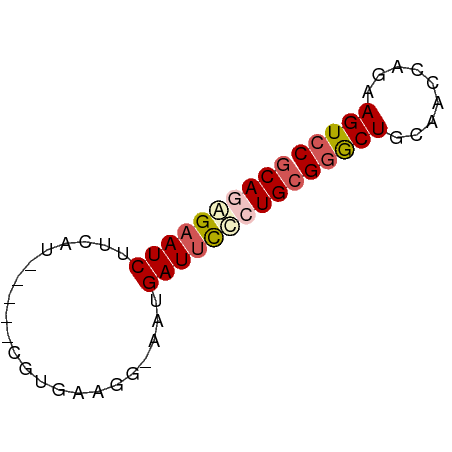
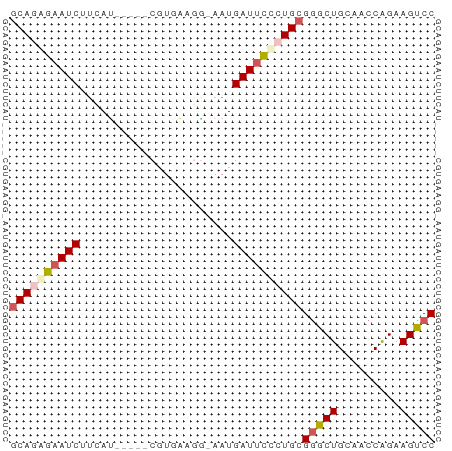
| Location | 5,684,823 – 5,684,878 |
|---|---|
| Length | 55 |
| Sequences | 5 |
| Columns | 61 |
| Reading direction | reverse |
| Mean pairwise identity | 75.26 |
| Shannon entropy | 0.41930 |
| G+C content | 0.48197 |
| Mean single sequence MFE | -14.92 |
| Consensus MFE | -8.59 |
| Energy contribution | -9.79 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.739949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5684823 55 - 22422827 GGACUUCUGGUUCCAGCCCGCAGAGAAUCAUU-CCUUCACG-----CUGAUGAUUCUCUGC ((....(((....))).))(((((((((((((-........-----..))))))))))))) ( -20.40, z-score = -3.33, R) >droSim1.chrX 4425310 55 - 17042790 GGACUUCUGGUUGCAGCCCGCAGGAUAUCAUU-ACUGCACG-----CUGAAGAUUCUCUGC (((..(((.....((((..((((.........-.))))..)-----))).)))...))).. ( -13.00, z-score = -0.09, R) >droSec1.super_4 5099861 61 + 6179234 GGACUUCUGGUUGCAGCCCGCAGGAAAUCAUUAUGAUUAAGAAAUCAUGAAGAUUCUCUGA ((.((.(.....).)).)).(((..((((.((((((((....)))))))).))))..))). ( -16.70, z-score = -1.72, R) >droYak2.chrX 18981891 55 + 21770863 GGACUCCCGAUUUCAGUACGCAUAGAAUCAUU-CCUUCACG-----AUUACGAUUCUCUGC ((....))...........(((.((((((..(-(......)-----)....)))))).))) ( -8.80, z-score = -0.82, R) >droEre2.scaffold_4690 3033434 55 - 18748788 GGACUUCUGGUUCCAGUCCGCAAGGAAUCAUU-CAUUCACG-----AUUCAGAUUCCGUGC (((((.........)))))(((.((((((..(-(......)-----)....)))))).))) ( -15.70, z-score = -2.12, R) >consensus GGACUUCUGGUUCCAGCCCGCAGGGAAUCAUU_CCUUCACG_____AUGAAGAUUCUCUGC ((....(((....))).))((((((((((......................)))))))))) ( -8.59 = -9.79 + 1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:18:35 2011