| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,680,528 – 5,680,640 |
| Length | 112 |
| Max. P | 0.642220 |

| Location | 5,680,528 – 5,680,640 |
|---|---|
| Length | 112 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.86 |
| Shannon entropy | 0.36280 |
| G+C content | 0.52415 |
| Mean single sequence MFE | -43.77 |
| Consensus MFE | -24.95 |
| Energy contribution | -26.51 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.642220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5680528 112 - 22422827 AAGGCGUCUUGGCCUGGCUGAAACGUGUUU-GUCUUGGCCAUU-----GGCCGAUGGACGCUUAGCCGCUUA-GUUGCGCUGCAAUUG-CCGUUUGAAACGAGUGCAAAAAGUGGUCAAA .(((((((((((((((((..(.(((....)-)).)..))))..-----)))))..)))))))).(((((((.-.(((((((((....)-)(((.....)))))))))).))))))).... ( -48.70, z-score = -3.77, R) >droAna3.scaffold_13117 1114006 117 - 5790199 AAGCCGGCUCUGUCGGGCUGGAACGUGUUU-GUCUUGGCCAUUGGCUCGGCUGAUGGCCGCUUAGCCGCUCA-GUGGCGCUGCAAUUG-CCGUUUGAAACGAGUGCAAAAAGUGGCCAAA .((((((((......(((..(.(((....)-)).)..)))...))).)))))..(((((((((.(((((...-)))))..((((....-.(((.....)))..))))..))))))))).. ( -47.10, z-score = -0.96, R) >droEre2.scaffold_4690 3029206 112 - 18748788 AAGGCGUCUUGGCCCGGCUAAAACGUGUUU-GUCUUGGCCAUU-----GGCCGAUGGCCGCUUAGCCGCUUA-GUUGCGCUGCAAUUG-CCGUUUGAAACGAGUGCAAAAAGUGGUCAAA .(((((((((((((.((((((.(((....)-)).))))))...-----)))))).)).))))).(((((((.-.(((((((((....)-)(((.....)))))))))).))))))).... ( -44.60, z-score = -2.18, R) >droYak2.chrX 18977665 112 + 21770863 AAGGCGUCUUGGCCUGGCUGAAACGUGUUU-GUCUUGGCCAUU-----GGCCGAUGGCCGCUUAGCCGCUUA-GUUGCGCUGCAAUUG-CCGUUUGAAACGAGUGCAAAAAGUGGUCAAA .(((((((((((((((((..(.(((....)-)).)..))))..-----)))))).)).))))).(((((((.-.(((((((((....)-)(((.....)))))))))).))))))).... ( -45.60, z-score = -2.51, R) >droSec1.super_4 5095556 112 + 6179234 AAGGCGUCUUGGCCUGGCUGAAACGUGUUU-GUCUUGGCCAUU-----GGCCGAUGGCCGCUUAGCCGCUUA-GUUGCGCUGCAAUUG-CCGUUUGAAACGAGUGCAAAAAGUGGUCAAA .(((((((((((((((((..(.(((....)-)).)..))))..-----)))))).)).))))).(((((((.-.(((((((((....)-)(((.....)))))))))).))))))).... ( -45.60, z-score = -2.51, R) >droSim1.chrX 4421172 111 - 17042790 AAGGCGUCUUGGCCUGGCUGAAACGUGUUU-GUCUUGGCCAUU-----GGCCGAUGGCCGCUUAGCCGCUUA-GUUGCGCUGCAA-UG-CCGUUUGAAACGAGUGCAAAAAGUGGUCAAA .(((((((((((((((((..(.(((....)-)).)..))))..-----)))))).)).))))).(((((((.-.(((((((((..-.)-)(((.....)))))))))).))))))).... ( -43.70, z-score = -1.91, R) >droVir3.scaffold_12932 1540809 120 + 2102469 GAGGCAGCUAGGCAAGUUGUUAACGUGUUUAGUUUUUGUGGCUAGCGAGCUUGCAGCGUGGAGCGUGGAGCGUGUAGCUUGGAGGUUGCCCAUUUGAAAUGAGUGCAAAAAAAAAACAAA ..(((((((...(((((((...((((.....((((((((.((.((....)).)).))).))))).....)))).)))))))..)))))))(((((.....)))))............... ( -31.10, z-score = 1.03, R) >consensus AAGGCGUCUUGGCCUGGCUGAAACGUGUUU_GUCUUGGCCAUU_____GGCCGAUGGCCGCUUAGCCGCUUA_GUUGCGCUGCAAUUG_CCGUUUGAAACGAGUGCAAAAAGUGGUCAAA .(((((((((((((.((((((.((.......)).))))))........)))))).)).))))).(((((((...(((((((.(((........))).....))))))).))))))).... (-24.95 = -26.51 + 1.56)


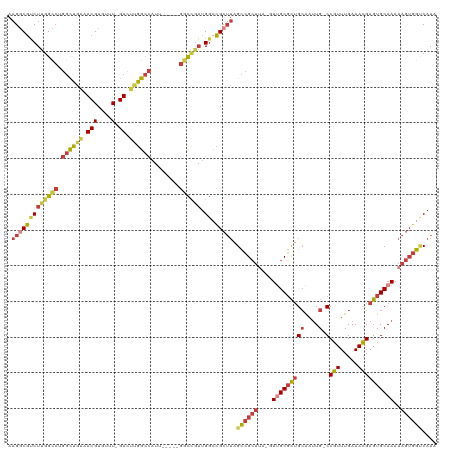
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:18:33 2011