| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,615,311 – 5,615,424 |
| Length | 113 |
| Max. P | 0.974489 |

| Location | 5,615,311 – 5,615,424 |
|---|---|
| Length | 113 |
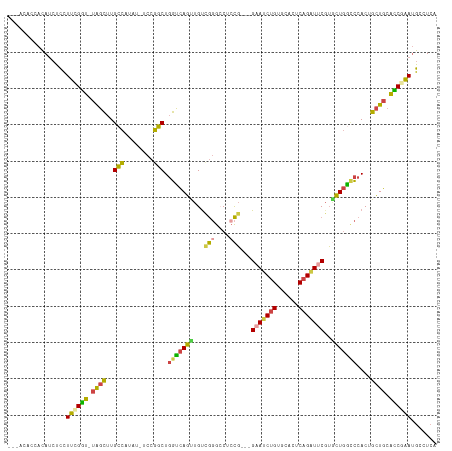
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 73.03 |
| Shannon entropy | 0.50844 |
| G+C content | 0.53387 |
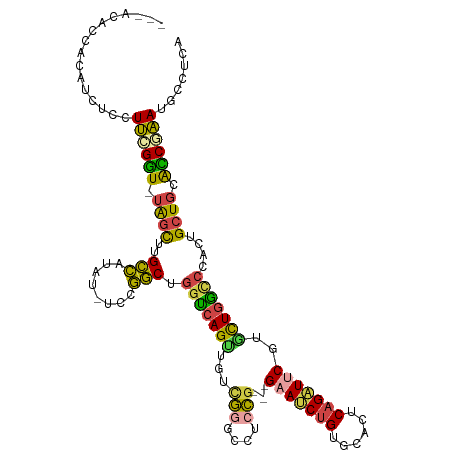
| Mean single sequence MFE | -34.95 |
| Consensus MFE | -25.62 |
| Energy contribution | -24.85 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.974489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5615311 113 - 22422827 ---AUAACACAUCUCCUUCGGU-UAGCUUGCCAUAU-UCCGGCUGGUCAGUUGUCGGGCCUCCGGAAGAAUCUGUGCACUCAGAUUCGUACUGGCCCACGGCUGCACCGAAUGCCUCA ---.............((((((-(((((.((((..(-(((((..((((........)))).))))))(((((((......)))))))....))))....))))).))))))....... ( -41.40, z-score = -2.77, R) >droSim1.chrX 4364821 110 - 17042790 ---ACACCACAUCUCCUUCGGU-UAGCUUGCCAUAU-UCCGGCUGGUCAGUUGUCGGGCCUCCG---GAAACUGUGCACUCAGAUUCGUGCUGGCCCACUGCUGCACCGAAUGCCUCA ---.............((((((-((((..((((..(-(((((..((((........)))).)))---))).....((((........)))))))).....)))).))))))....... ( -36.00, z-score = -1.23, R) >droSec1.super_4 5035478 110 + 6179234 ---ACACCACAUCUCCUUCGGU-UAGCUUGCCAUAU-UCCGGCUGGUCAGUUGUCGGGCCUCCG---GAAUCUGUGCACUCAGAUUCGUGCUGGCCCACUGCUGCACCGAAUGCCUCA ---................(((-......)))....-...(((((((((((.((.(((((..((---(((((((......))))))).))..))))))).)))).))))...)))... ( -39.10, z-score = -2.16, R) >droYak2.chrX 18916713 113 + 21770863 ACCACACCACAUCUCCUUUGGU-UAGCUUGCCAUAC-UCCGGCUGGUCAGUUGUUGGGGCUGCG---GAAUCUGAGGACUCAGAUUCGUGCUGGUCCACUGCUGCAUCUAAUGCCUCA ..................((((-......))))...-...(((.(((((((.((.(((.(.(((---((((((((....)))))))).))).).))))).)))).)))....)))... ( -35.00, z-score = -0.18, R) >droEre2.scaffold_4690 2968506 110 - 18748788 ---ACAACACAUCUCCUUCGGU-UAGCUUGCCAUAU-UCCGGCUGGUCAGCUGCCGGGGCUCCG---GUAUCUGAGGACUCAGAUUCGUGCUGACCCACUGCUGCACCUAAUGCCUCA ---................(((-((((..(((....-...))).(((((((((((((....)))---)))(((((....))))).....)))))))....)))).))).......... ( -37.30, z-score = -1.52, R) >droGri2.scaffold_14853 1734663 106 + 10151454 --------AUAACUUUUGUGUUGUGUUUUGUUGUGCGGUCAACUCAAAAGUCGU-AUGCCUCUUUUUGUAACCGGG---UCCGUUUCCGCUUCUGUAACCGUUUCGGCAUAAAUUUAA --------...(((((((.((((....(((.....))).)))).)))))))..(-(((((.............(((---......)))((....)).........))))))....... ( -20.90, z-score = -0.97, R) >consensus ___ACACCACAUCUCCUUCGGU_UAGCUUGCCAUAU_UCCGGCUGGUCAGUUGUCGGGCCUCCG___GAAUCUGUGCACUCAGAUUCGUGCUGGCCCACUGCUGCACCGAAUGCCUCA ................((((((.((((..(((........))).(((((((...(((....)))...(((((((......)))))))..)))))))....)))).))))))....... (-25.62 = -24.85 + -0.77)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:18:23 2011