
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,612,936 – 5,613,034 |
| Length | 98 |
| Max. P | 0.510662 |

| Location | 5,612,936 – 5,613,034 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 77.52 |
| Shannon entropy | 0.41010 |
| G+C content | 0.35588 |
| Mean single sequence MFE | -15.90 |
| Consensus MFE | -8.22 |
| Energy contribution | -8.92 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.510662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5612936 98 + 22422827 GGACUGAAUCACA-UAUUAGAGACGGUAAUAAACCGAUUUAGUGA-------AACACAAAA--AAAACAGGCAUAACCAA-----AUUUACAGCUUCUGAUAAA--CCCAGUACA ..((((.......-((((((((.((((.....)))).....(((.-------..)))....--......((.....))..-----.........))))))))..--..))))... ( -13.00, z-score = -0.92, R) >droSim1.chrX 4362554 93 + 17042790 -GACUGAAUCACA-UAUUAGAGACGGUAAUAAACCGAUUUAGUGA-------AACACAAAA--AAAACAGGCAUAA---------GUUUACAGCUUCUGAUAAA--CCCAGUACA -.((((.......-(((((((..((((.....)))).))))))).-------.........--.............---------((((((((...))).))))--).))))... ( -14.80, z-score = -1.73, R) >droSec1.super_4 5033198 92 - 6179234 -GACUGAAUCACA-UAUUAGAGACGGUAAUAAACCGAUUUAGUGA-------AACACAAAA--AAA-CAGGCAUAA---------GUUUACAGCUUCUGAUAAA--CCCAGUACA -.((((.......-(((((((..((((.....)))).))))))).-------.........--...-.........---------((((((((...))).))))--).))))... ( -14.80, z-score = -1.71, R) >droYak2.chrX 18914372 112 - 21770863 GCACUGAAUCACA-UAUUAGAGACGGUAAUAAACCGAUUUAGUGAUACACUAAACACAAAACAAAAACGGGCAUAACCAUUCCACGUUUACAGCUUCUGAUAAA--CCCACUACA .............-((((((((.((((.....)))).(((((((...)))))))..........(((((((.((....)).)).))))).....))))))))..--......... ( -18.70, z-score = -2.52, R) >droEre2.scaffold_4690 2966052 105 + 18748788 GCACUGAAUCACA-UAUUAGAGACGGUAAUAAACCGAUUUGGUGA-------AACACAAAACAAAAGAAGGCAUACCCAUUCCACGUUUACAGCUUCUGAUAAA--CCCAGCACA .(((..((((.(.-.....)....(((.....)))))))..))).-------..............(((((.....)).)))...((((((((...))).))))--)........ ( -15.30, z-score = -0.44, R) >droAna3.scaffold_13117 539007 112 - 5790199 AGACUGAAUCACAAUAUUCGAGGCGGUAAUAAACCGAUUUAAUGAUACA---AAAACAAAAAUAGCCUCAUAACCAUUCACAAUUUAUUACAGUCAGUGAUAAAAAUUCGCCACA .((((((((......))))((((((((.....))).(((....)))...---............)))))......................)))).((((.......)))).... ( -18.80, z-score = -2.59, R) >consensus GGACUGAAUCACA_UAUUAGAGACGGUAAUAAACCGAUUUAGUGA_______AACACAAAA__AAAACAGGCAUAACCA______GUUUACAGCUUCUGAUAAA__CCCAGUACA ..............((((((((.((((.....)))).....(((..........))).....................................))))))))............. ( -8.22 = -8.92 + 0.70)

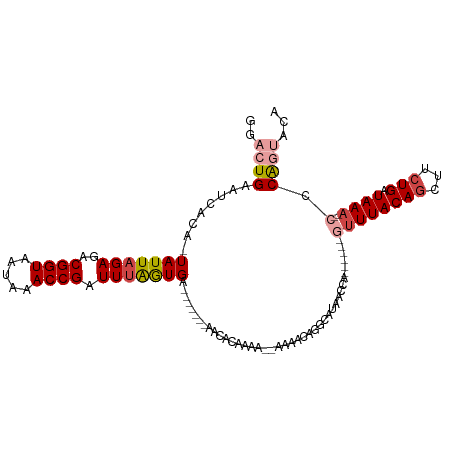

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:18:22 2011