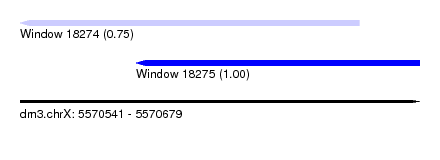
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,570,541 – 5,570,679 |
| Length | 138 |
| Max. P | 0.997714 |

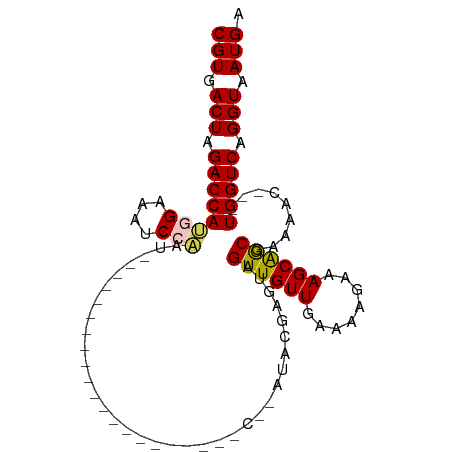
| Location | 5,570,541 – 5,570,658 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.46 |
| Shannon entropy | 0.38239 |
| G+C content | 0.39233 |
| Mean single sequence MFE | -23.40 |
| Consensus MFE | -16.32 |
| Energy contribution | -16.20 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.747882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5570541 117 - 22422827 CAAUGUAUUGCUAUUGGAUACAAUACAUAUACAAGUAGUUGUUGAAAAGAAAGCAGCGAAAAC---UGGUCAGGUAAUGAGUCAUCAUCAUCGGAGUCAUCUUGGCUGAUUAGCACAAAA ...(((((((.(((((....))))))).)))))....((((((........)))))).....(---(((((((.((((((.((..........)).))))..)).))))))))....... ( -27.10, z-score = -1.43, R) >droSim1.chrX 4321652 93 - 17042790 ------------------------CAAUAUACGAGUAGUUGUUGAAAAGAAAGCAGCGAAAAC---UGGUCAGGUAAUGAGUCAUCAUCAUCGGAGUCAUCUUGGCUGAUUAGCACAAAA ------------------------.............((((((........)))))).....(---(((((((.((((((.((..........)).))))..)).))))))))....... ( -20.00, z-score = -0.97, R) >droSec1.super_4 4991645 93 + 6179234 ------------------------CAAUAUACGAGUAGUUGUUGAAAAGAAAGCAGCGAAAAC---UGGUCAGGUAAUGAGUCAUCAUCAUCAGAGUCAUCUUGGCUGAUUAGCACAAAA ------------------------.............((((((........)))))).....(---(((((((.((((((.((..........)).))))..)).))))))))....... ( -20.10, z-score = -1.13, R) >droYak2.chrX 18872070 103 + 21770863 -----------CUAUAAUGGCUAUACGUAUACGUAUAGUUGUUGCAAAGAAAGCAGCGUAAACUGGUGGUCAGGUAAUGAGUCAUCA------GAGUCAUCUUGGCUGAUUAGCACAAAA -----------.......(((((((((....)))))))))(((((.......)))))......((.(((((((.((((((.((....------)).))))..)).))))))).))..... ( -31.20, z-score = -3.09, R) >droEre2.scaffold_4690 2923407 90 - 18748788 --------------------CAAAGCCAAUACAUAUAGUUGUUGAAAAGAAAGCGACGCAAAC---UGGUCAGGUAAUGCGUCAUC-------GAGUCAUCUUGGCUGAUUAGCACAAAA --------------------...((((((...........(((........)))((((((.((---(.....)))..))))))...-------........))))))............. ( -18.60, z-score = -0.40, R) >consensus ____________________C_A_ACAUAUACGAGUAGUUGUUGAAAAGAAAGCAGCGAAAAC___UGGUCAGGUAAUGAGUCAUCAUCAUC_GAGUCAUCUUGGCUGAUUAGCACAAAA .....................................((((((........)))))).........(((((((.((((((.((..........)).))))..)).)))))))........ (-16.32 = -16.20 + -0.12)

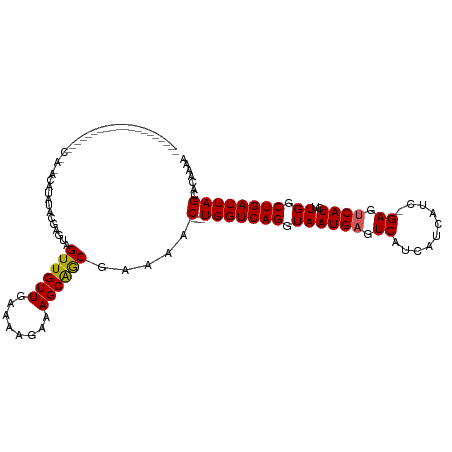
| Location | 5,570,581 – 5,570,679 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 76.76 |
| Shannon entropy | 0.37366 |
| G+C content | 0.39768 |
| Mean single sequence MFE | -21.46 |
| Consensus MFE | -16.88 |
| Energy contribution | -16.92 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.997714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5570581 98 - 22422827 CGUGACUAGACCAUGGAAAUCCAAUGUAUUGCUAUUGGAUACAAUACAUAUACAAGUAGUUGUUGAAAAGAAAGCAGCGAAAAC---UGGUCAGGUAAUGA (((.(((.((((((((....))).(((((((.(((((....))))))).)))))....((((((........))))))......---))))).))).))). ( -26.20, z-score = -3.24, R) >droSim1.chrX 4321692 74 - 17042790 CGUGACUAGACCAUGGAAAUCCAAU------------------------AUACGAGUAGUUGUUGAAAAGAAAGCAGCGAAAAC---UGGUCAGGUAAUGA (((.(((.((((((((....)))..------------------------.........((((((........))))))......---))))).))).))). ( -19.50, z-score = -3.39, R) >droSec1.super_4 4991685 74 + 6179234 CGUGACUAGACCAUGGAAAUCCAAU------------------------AUACGAGUAGUUGUUGAAAAGAAAGCAGCGAAAAC---UGGUCAGGUAAUGA (((.(((.((((((((....)))..------------------------.........((((((........))))))......---))))).))).))). ( -19.50, z-score = -3.39, R) >droYak2.chrX 18872104 90 + 21770863 CGUGACUAGACCAAUGAAAUCCUAU-----------AAUGGCUAUACGUAUACGUAUAGUUGUUGCAAAGAAAGCAGCGUAAACUGGUGGUCAGGUAAUGA (((.(((.(((((............-----------...(((((((((....)))))))))(((((.......))))).........))))).))).))). ( -27.30, z-score = -3.28, R) >droEre2.scaffold_4690 2923440 78 - 18748788 CGUGACUAGACCAAUGAAAUCCAAA--------------------GCCAAUACAUAUAGUUGUUGAAAAGAAAGCGACGCAAAC---UGGUCAGGUAAUGC (.(((((((.....((.....))..--------------------.............((((((........)))))).....)---)))))).)...... ( -14.80, z-score = -1.24, R) >consensus CGUGACUAGACCAUGGAAAUCCAAU_____________________C__AUACGAGUAGUUGUUGAAAAGAAAGCAGCGAAAAC___UGGUCAGGUAAUGA (((.(((.((((((((....)))...................................((((((........)))))).........))))).))).))). (-16.88 = -16.92 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:18:16 2011