| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,566,419 – 5,566,521 |
| Length | 102 |
| Max. P | 0.776200 |

| Location | 5,566,419 – 5,566,521 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 59.05 |
| Shannon entropy | 0.79154 |
| G+C content | 0.52053 |
| Mean single sequence MFE | -31.17 |
| Consensus MFE | -9.28 |
| Energy contribution | -7.91 |
| Covariance contribution | -1.37 |
| Combinations/Pair | 1.71 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.776200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
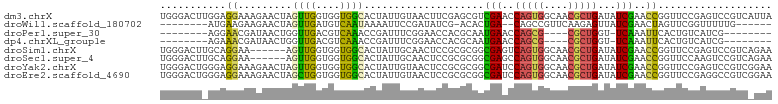
>dm3.chrX 5566419 102 - 22422827 UGGGACUUGGAGGAAAGAACUAGUUGGUGGUGGCACUAUUGUAACUUCGAGCGUCGAACCAGUGGCAACGCUGAUAUCGAACCGGUUCCGAGUCCGUCAUUA ..(((((((((((.........(((.(.(((.(((....))).))).).))).((((..(((((....)))))...)))).))...)))))))))....... ( -35.50, z-score = -1.82, R) >droWil1.scaffold_180702 811261 85 - 4511350 --------AUGAAGAAGAACUAGUUGAUGUCAAUAAAAUUCCGAUAUCG-ACACUGA--CAGCCGUUCAAGAGUUAUCGAACUAGUUCGGUUUUUG------ --------..(((((.(((((((((((((((...........)))))).-....(((--.((((......).))).))))))))))))..))))).------ ( -20.20, z-score = -1.65, R) >droPer1.super_30 318287 81 + 958394 --------AGGAACGAUAACUGGUUGACGUCAAACCGAUUUCGGAACCACGCAAUGAACCAGCG----CGCUGGU-UCAAAUUCACUGUCAUCG-------- --------.....((((.((.(((((.(((....(((....)))....)))))))(((((((..----..)))))-)).......).)).))))-------- ( -23.60, z-score = -1.87, R) >dp4.chrXL_group1e 572662 81 - 12523060 --------AGAAACGAUAACUGGUUGACGUCAAACCGAUUUCGGAACCACGCAAUGAACCAGCG----CGCUGGU-UCAAAUUCACUGUCAUCG-------- --------.....((((.((.(((((.(((....(((....)))....)))))))(((((((..----..)))))-)).......).)).))))-------- ( -23.60, z-score = -2.26, R) >droSim1.chrX 4315809 96 - 17042790 UGGGACUUGCAGGAA------AGUUGGUGGUGGCACUAUUGCAACUCCGCGCGGCGAGUCAGUGGCAACGCUGAUAUCGAACCGGUUCCGAGUCCGUCAGAA ..((((((...((((------.....((((..(((....)))....)))).(((((((((((((....))))))).)))..))).))))))))))....... ( -36.80, z-score = -1.44, R) >droSec1.super_4 4987591 96 + 6179234 UGGGACUUGCAGGAA------AGUUGGUGGUGGCACUAUUGCAACUCCGCGCGGCGAGCCAGUGGCAACGCUGAUAUCGAACCGGUUCCAAGUCCGUCAGAA ..(((((((..((..------.((((((((..(((....)))....)))).))))((..(((((....)))))...))...)).....)))))))....... ( -34.20, z-score = -0.65, R) >droYak2.chrX 18867875 102 + 21770863 UGGGACUGGGAGGAAAGAACUAGUUGGUGGUGGCACUAUUGUAACUCCGCGCGGCGAUCCAGUGGCAACGCUGAUAUCGAACCGGUUCCGAGUCCGUCGGAA ..(((((.((((..............((((.(..((....))..).)))).(((((((.(((((....)))))..))))..))).)))).)))))....... ( -37.30, z-score = -1.09, R) >droEre2.scaffold_4690 2919236 102 - 18748788 UGGGACUGGGAGGAAAGAACUAGCUGGUGGUGGCACUAUUGUAACUCCGCGCGGCGAUCCAGUGGCAACGCUGAUAUCGAACCGGUUCCGAGGCCGUCGGAA .((((((((.............((((((((.(..((....))..).)))).))))(((.(((((....)))))..)))...))))))))............. ( -38.20, z-score = -1.08, R) >consensus UGGGACU_GGAGGAAAGAACUAGUUGGUGGUGGCACUAUUGCAACUCCGCGCGGCGAACCAGUGGCAACGCUGAUAUCGAACCGGUUCCGAGUCCGUC_GAA ...........((.........((((....))))....................(((..(((((....)))))...)))..))................... ( -9.28 = -7.91 + -1.37)
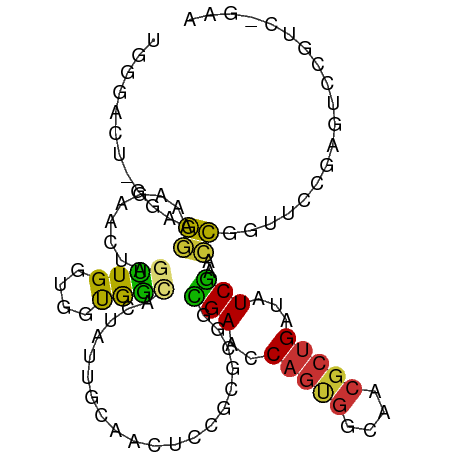
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:18:14 2011