
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,533,390 – 5,533,502 |
| Length | 112 |
| Max. P | 0.859382 |

| Location | 5,533,390 – 5,533,502 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 72.76 |
| Shannon entropy | 0.50210 |
| G+C content | 0.52419 |
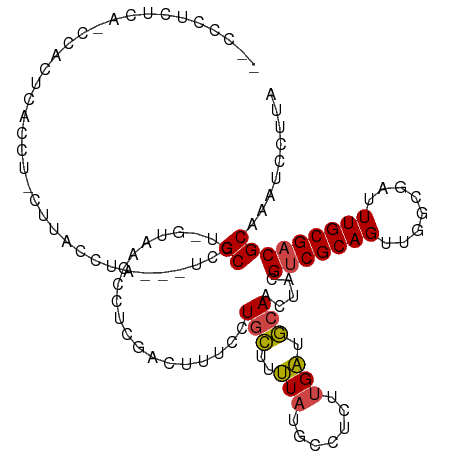
| Mean single sequence MFE | -20.35 |
| Consensus MFE | -11.45 |
| Energy contribution | -11.40 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.859382 |
| Prediction | RNA |
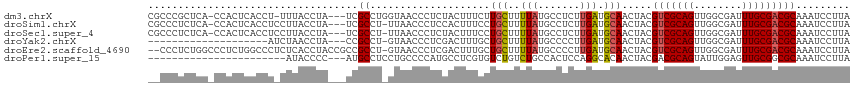
Download alignment: ClustalW | MAF
>dm3.chrX 5533390 112 - 22422827 CGCCCGCUCA-CCACUCACCU-UUUACCUA---UCGCCUGGUAACCCUCUACUUUCUUGCUUUUAUGCCUCUUGAUGCAACUACGUCGCAGUUGGCGAUUUGCGACGCAAAUCCUUA .(((.((((.-..........-.(((((..---......)))))..............((......)).....((((......))))).))).)))(((((((...))))))).... ( -21.70, z-score = -1.68, R) >droSim1.chrX 4282921 112 - 17042790 CGCCCUCUCA-CCACUCACCUCCUUACCUA---UCGCCU-UUAACCCUCCACUUUCCUGCUUUUAUGCCUCUUGAUGCAACUACGUCGCAGUUGGCGAUUUGCGACGCAAAUCCUUA .(((..(((.-...................---......-..................((......)).....((((......))))).))..)))(((((((...))))))).... ( -16.20, z-score = -1.62, R) >droSec1.super_4 4955278 112 + 6179234 CGCCCUCUCA-CCACUCACCUCCUUACCUA---UCGCCU-UUAACCCUCUACUUUCCUGCUUUUAUGCCUCUUGAUGCAACUACGUCGCAGUUGGCGAUUUGCGACGCAAAUCCUUA .(((..(((.-...................---......-..................((......)).....((((......))))).))..)))(((((((...))))))).... ( -16.20, z-score = -1.63, R) >droYak2.chrX 18834799 93 + 21770863 --------------------AUCUAACCUA---CCGCCU-GUAACCCUCGACUUUGCUGCUUUUAUGCCCCUUGAUGCAACUACGUCGCAGUUGGCGAUUUGCGACGCAAAUCCUUA --------------------..........---..(((.-..(((...((((...(.(((..(((.......))).))).)...))))..))))))(((((((...))))))).... ( -17.60, z-score = -1.23, R) >droEre2.scaffold_4690 2884467 114 - 18748788 --CCCUCUGGCCCUCUGGCCCUCUCACCUACCGCCGCCU-GUAACCCUCGACUUUGCUGCUUUUAUGCCCCUUGAUGCAACUACGUCGCAGUUGGCGAUUUGCGACGCAAAUCCUUA --......((((....))))............((((.((-((................((......)).....((((......)))))))).))))(((((((...))))))).... ( -27.20, z-score = -2.07, R) >droPer1.super_15 508164 91 - 2181545 -----------------------AUACCCC---AUGCCUCCUGCCCCAUGCCUCGUGUCUGUCUGCCACUCCAGGCACAACUACGACGCAGUAUUGGAGUUGCGGCGCAAAUCCUUA -----------------------.......---..(((.(..((.((((((.(((((..(((..(((......))))))..))))).)))....))).)).).)))........... ( -23.20, z-score = -0.61, R) >consensus __CCCUCUCA_CCACUCACCU_CUUACCUA___UCGCCU_GUAACCCUCGACUUUCCUGCUUUUAUGCCUCUUGAUGCAACUACGUCGCAGUUGGCGAUUUGCGACGCAAAUCCUUA ...................................((....................(((..(((.......))).))).....(((((((........)))))))))......... (-11.45 = -11.40 + -0.05)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:18:09 2011