| Sequence ID | dm3.chrX |
|---|---|
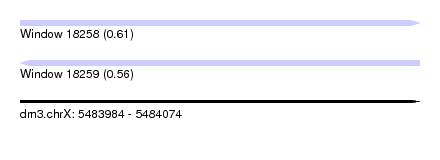
| Location | 5,483,984 – 5,484,074 |
| Length | 90 |
| Max. P | 0.611233 |

| Location | 5,483,984 – 5,484,074 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 74.53 |
| Shannon entropy | 0.52374 |
| G+C content | 0.37790 |
| Mean single sequence MFE | -17.19 |
| Consensus MFE | -7.57 |
| Energy contribution | -7.67 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.611233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5483984 90 + 22422827 ACCUCGCAGUGGGG--G------GAAAAA--------AAAACUGAGAA----CGAAAACACGUCAAUGCGAUAAUUAUUAAUAAUUAUCCACUCGUCCUUCUUUUGCUUC---- .....((((.((((--(------((....--------.....((((.(----((......)))...((.((((((((....))))))))))))))))))))).))))...---- ( -20.70, z-score = -2.19, R) >droSim1.chrX 4247277 91 + 17042790 CCCUCGCAGUGGGGA-A------GAAAAA--------AAAACUGAGAA----CGAAAACACGUCAAUGCGAUAAUUAUUAAUAAUUAUCCACUCGUCCUUCUUUUGCUUC---- .....((((.(((((-.------((....--------........((.----((......))))..((.((((((((....)))))))))).)).)))))...))))...---- ( -16.00, z-score = -0.91, R) >droSec1.super_4 4905357 92 - 6179234 CCCUCGCAGUGGGGAAG------AAAAAA--------AAAACUGAGAA----CGAAAACACGUCAAUGCGAUAAUUAUUAAUAAUUAUCCACUCGUCCUUCUUUUGCUUC---- .....((((.(((((.(------(.....--------........((.----((......))))..((.((((((((....)))))))))).)).)))))...))))...---- ( -16.00, z-score = -0.88, R) >droYak2.chrX 18786115 97 - 21770863 CCCUCGGACUGGAAAAAAA-UGAAAAAAA--------AAAACUGAGAA----CGAAAACACGUCAAUGCGAUAAUUAUUAAUAAUUAUCCACUCGUCCUUCUUUUGCUUC---- .....((((.((.......-.........--------........((.----((......))))..((.((((((((....)))))))))))).))))............---- ( -13.80, z-score = -1.31, R) >droEre2.scaffold_4690 2834952 98 + 18748788 CCCUCGUAGUGGAAGAAAAAUACGAAAAA--------AAAACUGAGAA----CGAAAACACGUCAAUGCGAUAAUUAUUAAUAAUUAUCCACUCGUCCUUCUUUUGCUUC---- .......((..((((((....((((....--------........((.----((......))))..((.((((((((....)))))))))).))))..))))))..))..---- ( -18.20, z-score = -2.39, R) >droAna3.scaffold_13117 4964225 106 - 5790199 CACUCACAUCAACAACAAAACACAAAAAAGACU---GAAAACUGAAAA----CGAAAACACGUCAAUGCGAUAAUUAUUAAUAAUUAUCCACUCGUCCUUCUUUUCCUUUUGC- .................................---.......(((((----.(((...(((....((.((((((((....))))))))))..)))..)))))))).......- ( -10.70, z-score = -1.51, R) >droPer1.super_15 1773665 110 - 2181545 CCUUUGCAAUGGCAGGGGGGGGGGGGGGGCACCCUCGAAGAGCGAGAAGAAGCGAAAACACGUCAAUGCGAUAAUUAUUAAUAAUUAUCCACUCGUCCUUCUUCUUUGGC---- (((((((....)))))))(((((((((....)))))((((..((((.....(((......)))...((.((((((((....))))))))))))))..))))..))))...---- ( -37.20, z-score = -2.00, R) >droWil1.scaffold_181096 11056953 85 + 12416693 ----------------AGUAGAAU--AAAU-------AAAACUGAAAA----CGCAAACACGUCAAUGCGAUAAUUAUUAAUAAUUAUCCACUCGUCCUUCUUCUGUUUCUCGG ----------------........--....-------....(((((((----((((..........)))((((((((....))))))))................)))).)))) ( -10.70, z-score = -1.05, R) >droVir3.scaffold_12928 5106457 92 + 7717345 CGCACACACAAAGGGCA-CAGAAU--AAAU-------AAAACUGAAAA----CGAAAACACGUCAAUGCGAUAAUUAUUAAUAAUUAUCCACUCGUCCUUCUUCUG-------- ..........((((((.-(((...--....-------....)))....----..............((.((((((((....))))))))))...))))))......-------- ( -13.50, z-score = -2.12, R) >droMoj3.scaffold_6359 3610493 95 + 4525533 CGCACACAGAAAGGCCAGCAGCAGCGAAAC-------AAAACUGAAAA----CGAAAACACGUCAAUGCGAUAAUUAUUAAUAAUUAUCCACUCGUCCUUCUUCUG-------- ......((((((((.(.(((...(((....-------...........----........)))...)))((((((((....)))))))).....).)))..)))))-------- ( -15.10, z-score = -1.10, R) >consensus CCCUCGCAGUGGGGA_A__AGAAGAAAAA________AAAACUGAGAA____CGAAAACACGUCAAUGCGAUAAUUAUUAAUAAUUAUCCACUCGUCCUUCUUUUGCUUC____ .....................................................(((...(((....((.((((((((....))))))))))..)))..)))............. ( -7.57 = -7.67 + 0.10)
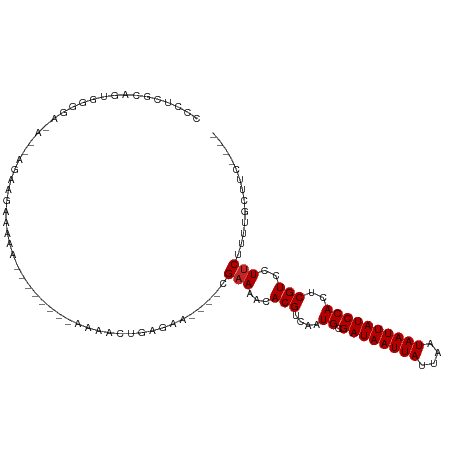

| Location | 5,483,984 – 5,484,074 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 74.53 |
| Shannon entropy | 0.52374 |
| G+C content | 0.37790 |
| Mean single sequence MFE | -20.30 |
| Consensus MFE | -11.97 |
| Energy contribution | -12.07 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.84 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.557375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
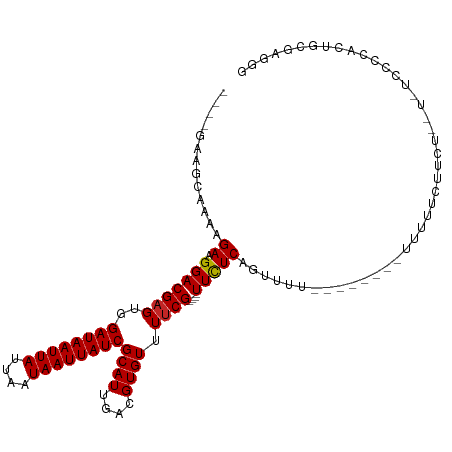
>dm3.chrX 5483984 90 - 22422827 ----GAAGCAAAAGAAGGACGAGUGGAUAAUUAUUAAUAAUUAUCGCAUUGACGUGUUUUCG----UUCUCAGUUUU--------UUUUUC------C--CCCCACUGCGAGGU ----...(((((((((((((.((((((((((((....)))))))).))))((((......))----))....)))))--------))))).------.--......)))..... ( -17.90, z-score = -0.49, R) >droSim1.chrX 4247277 91 - 17042790 ----GAAGCAAAAGAAGGACGAGUGGAUAAUUAUUAAUAAUUAUCGCAUUGACGUGUUUUCG----UUCUCAGUUUU--------UUUUUC------U-UCCCCACUGCGAGGG ----((((..((((((((((.((((((((((((....)))))))).))))((((......))----))....)))))--------))))))------)-))(((.......))) ( -21.60, z-score = -1.49, R) >droSec1.super_4 4905357 92 + 6179234 ----GAAGCAAAAGAAGGACGAGUGGAUAAUUAUUAAUAAUUAUCGCAUUGACGUGUUUUCG----UUCUCAGUUUU--------UUUUUU------CUUCCCCACUGCGAGGG ----((((.(((((((((((.((((((((((((....)))))))).))))((((......))----))....)))))--------))))))------))))(((.......))) ( -24.40, z-score = -2.41, R) >droYak2.chrX 18786115 97 + 21770863 ----GAAGCAAAAGAAGGACGAGUGGAUAAUUAUUAAUAAUUAUCGCAUUGACGUGUUUUCG----UUCUCAGUUUU--------UUUUUUUCA-UUUUUUUCCAGUCCGAGGG ----...(.(((((((((((.((((((((((((....)))))))).))))((((......))----))....)))))--------)))))).).-.......((.......)). ( -18.20, z-score = -0.65, R) >droEre2.scaffold_4690 2834952 98 - 18748788 ----GAAGCAAAAGAAGGACGAGUGGAUAAUUAUUAAUAAUUAUCGCAUUGACGUGUUUUCG----UUCUCAGUUUU--------UUUUUCGUAUUUUUCUUCCACUACGAGGG ----((((.(((((.((((((((..((((((((....))))))))((((....)))).))))----))))...))))--------).))))........((((......)))). ( -20.20, z-score = -1.07, R) >droAna3.scaffold_13117 4964225 106 + 5790199 -GCAAAAGGAAAAGAAGGACGAGUGGAUAAUUAUUAAUAAUUAUCGCAUUGACGUGUUUUCG----UUUUCAGUUUUC---AGUCUUUUUUGUGUUUUGUUGUUGAUGUGAGUG -((((((.(..(((((((((.((((((((((((....)))))))).))))((((......))----))..........---.))))))))).).)))))).............. ( -22.10, z-score = -1.00, R) >droPer1.super_15 1773665 110 + 2181545 ----GCCAAAGAAGAAGGACGAGUGGAUAAUUAUUAAUAAUUAUCGCAUUGACGUGUUUUCGCUUCUUCUCGCUCUUCGAGGGUGCCCCCCCCCCCCCCCUGCCAUUGCAAAGG ----(((...(((((((((.(((((((((((((....))))))))((((....))))...))))).))))...)))))...)))............((..(((....)))..)) ( -25.40, z-score = -0.48, R) >droWil1.scaffold_181096 11056953 85 - 12416693 CCGAGAAACAGAAGAAGGACGAGUGGAUAAUUAUUAAUAAUUAUCGCAUUGACGUGUUUGCG----UUUUCAGUUUU-------AUUU--AUUCUACU---------------- ....(((((.(((((.(((((((((((((((((....)))))))).))))....)))))...----))))).)))))-------....--........---------------- ( -16.10, z-score = -0.89, R) >droVir3.scaffold_12928 5106457 92 - 7717345 --------CAGAAGAAGGACGAGUGGAUAAUUAUUAAUAAUUAUCGCAUUGACGUGUUUUCG----UUUUCAGUUUU-------AUUU--AUUCUG-UGCCCUUUGUGUGUGCG --------(((((..(((((.((((((((((((....)))))))).))))((((......))----))....)))))-------....--.)))))-.((.(.......).)). ( -16.40, z-score = 0.00, R) >droMoj3.scaffold_6359 3610493 95 - 4525533 --------CAGAAGAAGGACGAGUGGAUAAUUAUUAAUAAUUAUCGCAUUGACGUGUUUUCG----UUUUCAGUUUU-------GUUUCGCUGCUGCUGGCCUUUCUGUGUGCG --------(((((((((((((((((((((((((....)))))))).))))..))).))))).----...(((((...-------((......)).)))))...)))))...... ( -20.70, z-score = 0.09, R) >consensus ____GAAGCAAAAGAAGGACGAGUGGAUAAUUAUUAAUAAUUAUCGCAUUGACGUGUUUUCG____UUCUCAGUUUU________UUUUUCUUCU__U_UCCCCACUGCGAGGG .............((((((((((((((((((((....)))))))).))))..))).)))))..................................................... (-11.97 = -12.07 + 0.10)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:18:03 2011