| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,439,146 – 5,439,265 |
| Length | 119 |
| Max. P | 0.904025 |

| Location | 5,439,146 – 5,439,265 |
|---|---|
| Length | 119 |
| Sequences | 8 |
| Columns | 123 |
| Reading direction | forward |
| Mean pairwise identity | 73.30 |
| Shannon entropy | 0.51756 |
| G+C content | 0.47430 |
| Mean single sequence MFE | -25.14 |
| Consensus MFE | -10.97 |
| Energy contribution | -11.41 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.904025 |
| Prediction | RNA |
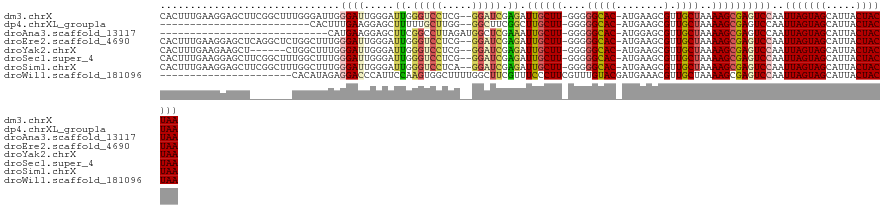
Download alignment: ClustalW | MAF
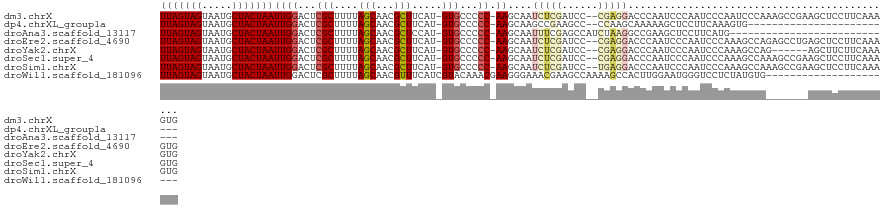
>dm3.chrX 5439146 119 + 22422827 UUAGUAGUAAUGCUACUAAUUGGACUCGCUUUUAGCAACGCUUCAU-GUGCCCCC-AAGCAAUCUCGAUCC--CGAGGACCCAAUCCCAAUCCCAAUCCCAAAGCCGAAGCUCCUUCAAAGUG (((((((.....)))))))..(((...((.....))...(((((..-.(((....-..))).(((((....--)))))............................))))))))......... ( -24.70, z-score = -2.42, R) >dp4.chrXL_group1a 1049054 94 - 9151740 UUAGUAGUAAUGCUACUAAUUGGACUCGCUUUUAGCAACGCUUCAU-GUGCCCCC-AAGCAAGCCGAAGCC--CCAAGCAAAAAGCUCCUUCAAAGUG------------------------- (((((((.....)))))))(((((...((((((.((...(((((..-.(((....-..)))....))))).--....)).))))))...)))))....------------------------- ( -25.10, z-score = -2.75, R) >droAna3.scaffold_13117 4919559 93 - 5790199 UUAGUAGUAAUGCUACUAAUUGGACUCGCUUUUAGCAACGCUCCAU-GUGCCCCC-AAGCAAUUUCGAGCCAUCUAAGGCCGAAGCUCCUUCAUG---------------------------- (((((((.....)))))))..(((((((.((((((....((((...-.(((....-..))).....))))...)))))).)).)).)))......---------------------------- ( -23.10, z-score = -1.77, R) >droEre2.scaffold_4690 2789418 119 + 18748788 UUAGUAGUAAUGCUACUAAUUGGACUCGCUUUUAGCAACGCUUCAU-GUGCCCCC-AAGCAAUCUCGAUCC--CGAGGACCCAAUCCCAAUCCCAAAGCCAGAGCCUGAGCUCCUUCAAAGUG (((((((.....)))))))..(((((((((((..((...((((...-........-))))..(((((....--)))))...................)).)))))..))).)))......... ( -25.20, z-score = -1.35, R) >droYak2.chrX 18740672 113 - 21770863 UUAGUAGUAAUGCUACUAAUUGGACUCGCUUUUAGCAACGCUUCAU-GUGCCCCC-AAGCAAUCUCGAUCC--CGAGGACCCAAUCCCAAUCCCAAAGCCAG------AGCUUCUUCAAAGUG (((((((.....)))))))(((((...(((((..((...((((...-........-))))..(((((....--)))))...................)).))------)))...))))).... ( -21.90, z-score = -1.06, R) >droSec1.super_4 4862024 119 - 6179234 UUAGUAGUAAUGCUACUAAUUGGACUCGCUUUUAGCAACGCUUCAU-GUGCCCCC-AAGCAAUCUCGAUCC--CGAGGACCCAAUCCCAAUCCCAAAGCCAAAGCCGAAGCUCCUUCAAAGUG (((((((.....)))))))..(((...((.....))...(((((..-.(((....-..))).(((((....--)))))............................))))))))......... ( -24.70, z-score = -1.88, R) >droSim1.chrX 4203503 119 + 17042790 UUAGUAGUAAUGCUACUAAUUGGACUCGCUUUUAGCAACGCUUCAU-GUGCCCCC-AAGCAAUCUCGAUCC--UGAGGACCCAAUCCCAAUCCCAAAGCCAAAGCCGAAGCUCCUUCAAAGUG (((((((.....)))))))..(((...((.....))...(((((..-.(((....-..))).(((((....--)))))............................))))))))......... ( -22.70, z-score = -0.96, R) >droWil1.scaffold_181096 10981915 101 + 12416693 UUAGUAGUAAUGCUACUAAUUGGACUCGCUUUUAGCAACGUUUCAUCGUACAAACGAAGGGAAACGAAGCCAAAAGCCACUUGGAAUGGGUCCUCUAUGUG---------------------- (((((((.....)))))))..((((((((((((.((..((((((.((((....))))...))))))..)).))))))(....)....))))))........---------------------- ( -33.70, z-score = -4.04, R) >consensus UUAGUAGUAAUGCUACUAAUUGGACUCGCUUUUAGCAACGCUUCAU_GUGCCCCC_AAGCAAUCUCGAUCC__CGAGGACCCAAUCCCAAUCCCAAAGCCA_______AGCUCCUUCAAAGUG (((((((.....)))))))((((....((((........((........)).....))))..(((((......)))))..))))....................................... (-10.97 = -11.41 + 0.44)
| Location | 5,439,146 – 5,439,265 |
|---|---|
| Length | 119 |
| Sequences | 8 |
| Columns | 123 |
| Reading direction | reverse |
| Mean pairwise identity | 73.30 |
| Shannon entropy | 0.51756 |
| G+C content | 0.47430 |
| Mean single sequence MFE | -32.56 |
| Consensus MFE | -17.18 |
| Energy contribution | -17.62 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.571185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
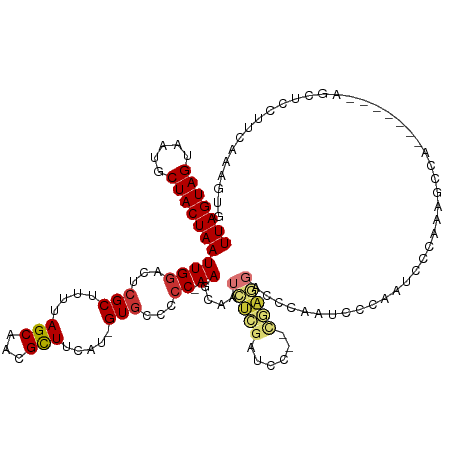
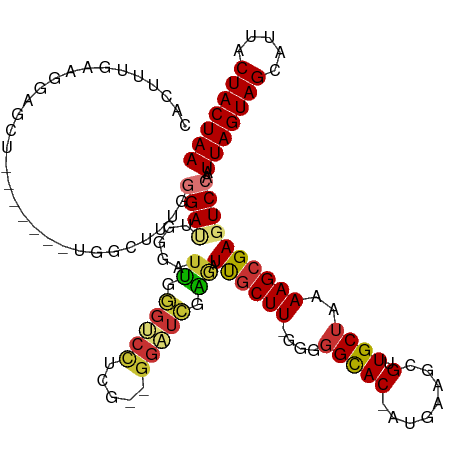
>dm3.chrX 5439146 119 - 22422827 CACUUUGAAGGAGCUUCGGCUUUGGGAUUGGGAUUGGGAUUGGGUCCUCG--GGAUCGAGAUUGCUU-GGGGGCAC-AUGAAGCGUUGCUAAAAGCGAGUCCAAUUAGUAGCAUUACUACUAA ..((.....(((((....)))))......))(((((((.((.(((((...--))))).)).((((((-...((((.-(((...)))))))..)))))).)))))))(((((.....))))).. ( -33.10, z-score = -0.31, R) >dp4.chrXL_group1a 1049054 94 + 9151740 -------------------------CACUUUGAAGGAGCUUUUUGCUUGG--GGCUUCGGCUUGCUU-GGGGGCAC-AUGAAGCGUUGCUAAAAGCGAGUCCAAUUAGUAGCAUUACUACUAA -------------------------....(((..((((((((......))--))))))(((((((((-...((((.-(((...)))))))..))))))))))))(((((((.....))))))) ( -30.60, z-score = -1.55, R) >droAna3.scaffold_13117 4919559 93 + 5790199 ----------------------------CAUGAAGGAGCUUCGGCCUUAGAUGGCUCGAAAUUGCUU-GGGGGCAC-AUGGAGCGUUGCUAAAAGCGAGUCCAAUUAGUAGCAUUACUACUAA ----------------------------......((((((((.(((((....(((........))).-.)))))..-..))))).((((.....)))).)))..(((((((.....))))))) ( -28.00, z-score = -1.17, R) >droEre2.scaffold_4690 2789418 119 - 18748788 CACUUUGAAGGAGCUCAGGCUCUGGCUUUGGGAUUGGGAUUGGGUCCUCG--GGAUCGAGAUUGCUU-GGGGGCAC-AUGAAGCGUUGCUAAAAGCGAGUCCAAUUAGUAGCAUUACUACUAA ..((..(..(((((....)))))..)...))(((((((.((.(((((...--))))).)).((((((-...((((.-(((...)))))))..)))))).)))))))(((((.....))))).. ( -34.20, z-score = 0.05, R) >droYak2.chrX 18740672 113 + 21770863 CACUUUGAAGAAGCU------CUGGCUUUGGGAUUGGGAUUGGGUCCUCG--GGAUCGAGAUUGCUU-GGGGGCAC-AUGAAGCGUUGCUAAAAGCGAGUCCAAUUAGUAGCAUUACUACUAA .........(((((.------...)))))..(((((((.((.(((((...--))))).)).((((((-...((((.-(((...)))))))..)))))).)))))))(((((.....))))).. ( -29.80, z-score = 0.25, R) >droSec1.super_4 4862024 119 + 6179234 CACUUUGAAGGAGCUUCGGCUUUGGCUUUGGGAUUGGGAUUGGGUCCUCG--GGAUCGAGAUUGCUU-GGGGGCAC-AUGAAGCGUUGCUAAAAGCGAGUCCAAUUAGUAGCAUUACUACUAA ..((..(..(((((....)))))..)...))(((((((.((.(((((...--))))).)).((((((-...((((.-(((...)))))))..)))))).)))))))(((((.....))))).. ( -34.50, z-score = -0.44, R) >droSim1.chrX 4203503 119 - 17042790 CACUUUGAAGGAGCUUCGGCUUUGGCUUUGGGAUUGGGAUUGGGUCCUCA--GGAUCGAGAUUGCUU-GGGGGCAC-AUGAAGCGUUGCUAAAAGCGAGUCCAAUUAGUAGCAUUACUACUAA ..((..(..(((((....)))))..)...))(((((((..((.(((((((--((..........)))-)))))).)-).....((((......))))..)))))))(((((.....))))).. ( -34.70, z-score = -0.54, R) >droWil1.scaffold_181096 10981915 101 - 12416693 ----------------------CACAUAGAGGACCCAUUCCAAGUGGCUUUUGGCUUCGUUUCCCUUCGUUUGUACGAUGAAACGUUGCUAAAAGCGAGUCCAAUUAGUAGCAUUACUACUAA ----------------------........((((.(((.....)))(((((((((..((((((...((((....)))).))))))..)))))))))..))))..(((((((.....))))))) ( -35.60, z-score = -5.24, R) >consensus CACUUUGAAGGAGCU_______UGGCUUUGGGAUUGGGAUUGGGUCCUCG__GGAUCGAGAUUGCUU_GGGGGCAC_AUGAAGCGUUGCUAAAAGCGAGUCCAAUUAGUAGCAUUACUACUAA ..............................((((.....((.(((((.....))))).)).((((((....((((...........))))..))))))))))..(((((((.....))))))) (-17.18 = -17.62 + 0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:18:00 2011