| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,419,186 – 5,419,287 |
| Length | 101 |
| Max. P | 0.998232 |

| Location | 5,419,186 – 5,419,287 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 59.27 |
| Shannon entropy | 0.70908 |
| G+C content | 0.51270 |
| Mean single sequence MFE | -31.33 |
| Consensus MFE | -19.92 |
| Energy contribution | -19.92 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.29 |
| SVM RNA-class probability | 0.998232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
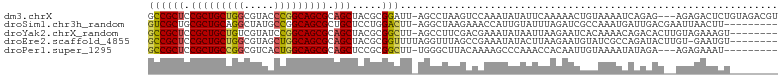
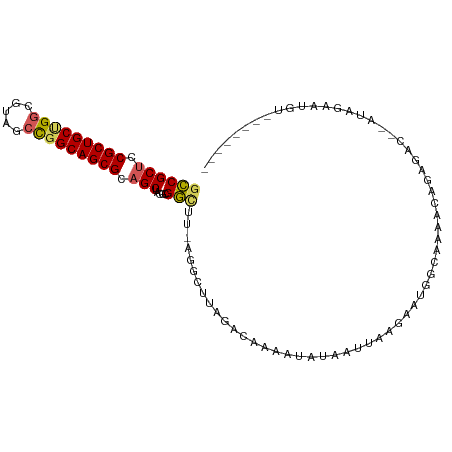
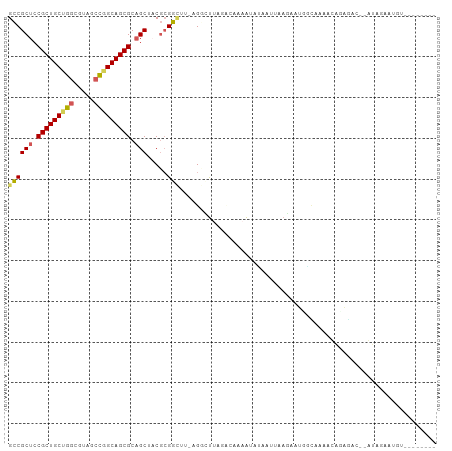
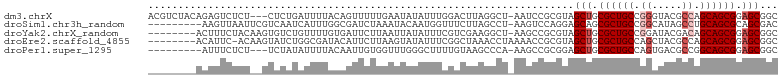
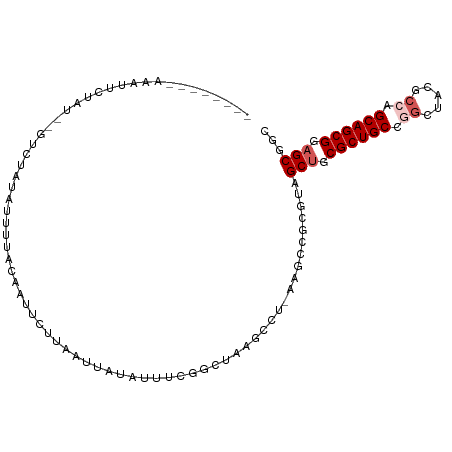
>dm3.chrX 5419186 101 + 22422827 GCCGCUCCGCUGCUGGCGUACCCGGCAGCGCAGCUACGCGGAUU-AGCCUAAGUCCAAAUAUAUUCAAAAACUGUAAAAUCAGAG---AGAGACUCUGUAGACGU ((.(((.(((((((((.....))))))))).)))...))(((((-......))))).................((.....(((((---.....)))))...)).. ( -32.80, z-score = -2.14, R) >droSim1.chr3h_random 862078 95 + 1452968 GUCGCUGCGCUGCAGGCUAUGCCGGCAGCGCUGCUCCUGGACUU-AGGCUAAGAAACCAUUGUAUUUAGAUCGCCAAAUGAUUGACGAAUUAACUU--------- (((((.(((((((.(((...))).))))))).))..........-.(((.......................)))........)))..........--------- ( -27.90, z-score = -0.78, R) >droYak2.chrX_random 1263068 96 - 1802292 GCCGCUCCGCUGCUGUCGUAUCCGGCAGCGCAGCUACGCGGCUU-AGCCUUCGACGAAAUAUAAUUAAGAAUCACAAAACAGACACUUGUAGAAAGU-------- (((((...((((((((((....)))))..)))))...)))))..-.((.(((.((((.............................)))).))).))-------- ( -24.45, z-score = -0.32, R) >droEre2.scaffold_4855 234991 96 - 242605 GCCGCUCCGCUGCUGGCGUAGCUGGCAGCGCAGCUACGCGGUUUUAGGUUUAGCCGAAAUAUACUUAAGAAUGUAUCGCCAGAUACUUGU-GAAUGU-------- ...(((.((((((..((...))..)))))).)))..(((((((((((((.((.......)).))))))))..(((((....)))))))))-).....-------- ( -33.50, z-score = -1.06, R) >droPer1.super_1295 3585 92 + 6930 GCCGCUCCGCUGCCGGCGUCACUGGCAGCGCAGCUCCGCGGCUU-UGGGCUUACAAAAGCCCAAACCACAAUUGUAAAAUAUAGA---AGAGAAAU--------- ((.(((.(((((((((.....))))))))).)))...))((.((-(((((((....)))))))))))..................---........--------- ( -38.00, z-score = -3.69, R) >consensus GCCGCUCCGCUGCUGGCGUAGCCGGCAGCGCAGCUACGCGGCUU_AGGCUUAGACAAAAUAUAAUUAAGAAUGGCAAAACAGAGAC__AUAGAAUGU________ ((((((.(((((((((.....))))))))).))).....)))............................................................... (-19.92 = -19.92 + -0.00)
| Location | 5,419,186 – 5,419,287 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 59.27 |
| Shannon entropy | 0.70908 |
| G+C content | 0.51270 |
| Mean single sequence MFE | -32.16 |
| Consensus MFE | -16.88 |
| Energy contribution | -17.68 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.986718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5419186 101 - 22422827 ACGUCUACAGAGUCUCU---CUCUGAUUUUACAGUUUUUGAAUAUAUUUGGACUUAGGCU-AAUCCGCGUAGCUGCGCUGCCGGGUACGCCAGCAGCGGAGCGGC ..((((((((((.....---)))))((((..........)))).....))))).((.((.-.....)).))(((.((((((.((.....)).)))))).)))... ( -31.90, z-score = -0.68, R) >droSim1.chr3h_random 862078 95 - 1452968 ---------AAGUUAAUUCGUCAAUCAUUUGGCGAUCUAAAUACAAUGGUUUCUUAGCCU-AAGUCCAGGAGCAGCGCUGCCGGCAUAGCCUGCAGCGCAGCGAC ---------...(((..(((((((....)))))))..)))........((((((..((..-..))..)))))).(((((((.(((...))).)))))))...... ( -33.10, z-score = -2.62, R) >droYak2.chrX_random 1263068 96 + 1802292 --------ACUUUCUACAAGUGUCUGUUUUGUGAUUCUUAAUUAUAUUUCGUCGAAGGCU-AAGCCGCGUAGCUGCGCUGCCGGAUACGACAGCAGCGGAGCGGC --------.(((((.((((((((..(((..(.....)..))).)))))).)).)))))..-..(((((....((((((((.((....)).)))).)))).))))) ( -30.20, z-score = -0.66, R) >droEre2.scaffold_4855 234991 96 + 242605 --------ACAUUC-ACAAGUAUCUGGCGAUACAUUCUUAAGUAUAUUUCGGCUAAACCUAAAACCGCGUAGCUGCGCUGCCAGCUACGCCAGCAGCGGAGCGGC --------......-...........(((((((........)))).....((.....))......)))...(((.((((((..((...))..)))))).)))... ( -24.10, z-score = 0.58, R) >droPer1.super_1295 3585 92 - 6930 ---------AUUUCUCU---UCUAUAUUUUACAAUUGUGGUUUGGGCUUUUGUAAGCCCA-AAGCCGCGGAGCUGCGCUGCCAGUGACGCCGGCAGCGGAGCGGC ---------........---..............(((((((((((((((....)))))).-))))))))).(((.(((((((.(.....).))))))).)))... ( -41.50, z-score = -3.40, R) >consensus ________AAAUUCUAU__GUCUAUAUUUUACAAUUCUUAAUUAUAUUUCGGCUAAGCCU_AAGCCGCGUAGCUGCGCUGCCGGCUACGCCAGCAGCGGAGCGGC .......................................................................(((.((((((.((.....)).)))))).)))... (-16.88 = -17.68 + 0.80)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:17:59 2011