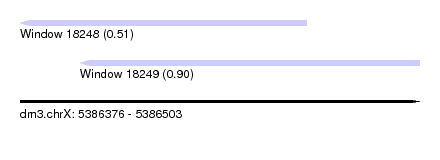
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,386,376 – 5,386,503 |
| Length | 127 |
| Max. P | 0.898985 |

| Location | 5,386,376 – 5,386,467 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 76.69 |
| Shannon entropy | 0.43646 |
| G+C content | 0.60223 |
| Mean single sequence MFE | -36.73 |
| Consensus MFE | -23.20 |
| Energy contribution | -23.07 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.506531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
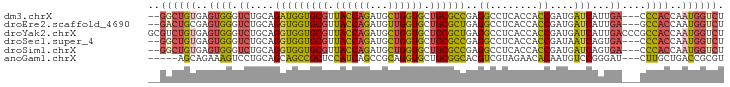
>dm3.chrX 5386376 91 - 22422827 --GGCUGUGAGUGGGUCUGCAGAUGGUGCGUUACCAGAUGCUGGUGCUGCGCCGAGGCCUCACCACCGAUGAUCAUUGA---CCCACCAAUGGUCU --..(.(((.((((((((.....(((((((.((((((...)))))).)))))))))))).)))))).)..((((((((.---.....)))))))). ( -38.40, z-score = -1.90, R) >droEre2.scaffold_4690 2738727 91 - 18748788 --GACUGCGAGUGGGUCUGCAGGUGGUGCGUUACCAGAUGUUGGUGCUGCGCUGAGGCCUCACCACCGAUGAUCAUUGA---GCCACCAAUGGUCU --.....((.((((....((...(((((((.((((((...)))))).)))))))..))....))))))..((((((((.---.....)))))))). ( -32.80, z-score = -0.24, R) >droYak2.chrX 18689253 96 + 21770863 GCGUCUGUGAGUGGGUCUGCAGGUGGUGCGUUACCAGAUGCUGGUGCUGCGCUGAGGCCUCACCACCGAUGAUCAUUGACCCGCCACCAAUGGUCU .(((..(((.(((((((....(((((((((.((((((...)))))).)))((....))...))))))(.....)...))))))))))..))).... ( -38.10, z-score = -0.84, R) >droSec1.super_4 4803885 91 + 6179234 --GGCUGUGAGUGGGUCUGCAGGUGGUGCGUUACCAGAUGCUGGUGCUGCGCCGAGGCCUCACCACCGAUAAUCAGUGA---CCCACCAAUGGUCU --((((((..(((((((.((.(((((((((.((((((...)))))).)))((....))...))))))(.....).))))---)))))..)))))). ( -41.40, z-score = -2.26, R) >droSim1.chrX 4154904 91 - 17042790 --GGCUGUGAGUGGGUCUGCAGGUGGUGCGUUACCAGAUGCUGGUGCUGCGCCGAGGCCUCACCACCGAUGAUCAGUGA---CCCACCAAUGGUCU --((((((..(((((((.((.(((((((((.((((((...)))))).)))((....))...))))))(.....).))))---)))))..)))))). ( -41.40, z-score = -1.93, R) >anoGam1.chrX 12328881 88 + 22145176 -----AGCAGAAAGUCCUGCAGCAGCCGCUCCAUCAGCCGCAGGUGCUGCGGCACGUCGUAGAACACAAUGUCCGGGAU---CUUGCUGACCGCGU -----..(((.(((((((((((((((.(((.....))).))...))))))((.((((.((.....)).)))))))))).---))).)))....... ( -28.30, z-score = 0.65, R) >consensus __GGCUGUGAGUGGGUCUGCAGGUGGUGCGUUACCAGAUGCUGGUGCUGCGCCGAGGCCUCACCACCGAUGAUCAGUGA___CCCACCAAUGGUCU ..((((((..((((..........((((((.((((((...)))))).))))))..((........))................))))..)))))). (-23.20 = -23.07 + -0.13)

| Location | 5,386,395 – 5,386,503 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 81.36 |
| Shannon entropy | 0.36288 |
| G+C content | 0.62260 |
| Mean single sequence MFE | -45.18 |
| Consensus MFE | -34.73 |
| Energy contribution | -34.82 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.898985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
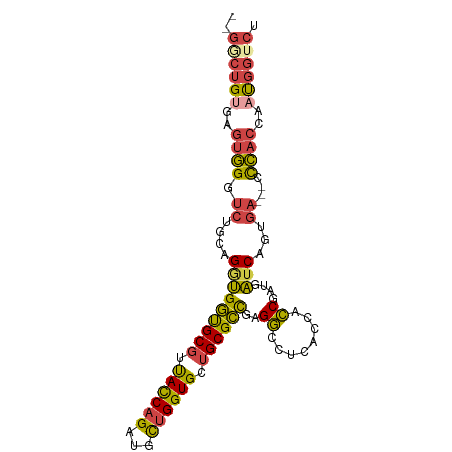
>dm3.chrX 5386395 108 - 22422827 AUGAUGUCCAUGGUGCUGACCCGCCGCUGCUGACUG---GGCUGUGAGUGGGUCUGCAGAUGGUGCGUUACCAGAUGCUGGUGCUGCGCCGAGGCCUCACCACCGAUGAUC ..((((((..(((((..((((((((((.(((.....---))).))).))))))).((...(((((((.((((((...)))))).)))))))..))..)))))..))).))) ( -46.60, z-score = -1.92, R) >droEre2.scaffold_4690 2738746 108 - 18748788 AUGAUGUCCAUGGUGCUGACCCGCCGCUGCUGACUG---GACUGCGAGUGGGUCUGCAGGUGGUGCGUUACCAGAUGUUGGUGCUGCGCUGAGGCCUCACCACCGAUGAUC ..((((((..((((((.((((((((((..((....)---)...))).))))))).))((((((((((.((((((...)))))).))))))...)))).))))..))).))) ( -43.50, z-score = -1.23, R) >droYak2.chrX 18689275 111 + 21770863 AUGAUGUCCAUGGUGCUGACCCGCCGCUGCUGACUGUGCGUCUGUGAGUGGGUCUGCAGGUGGUGCGUUACCAGAUGCUGGUGCUGCGCUGAGGCCUCACCACCGAUGAUC ..((((((..((((((.((((((((((....(((.....))).))).))))))).))((((((((((.((((((...)))))).))))))...)))).))))..))).))) ( -45.40, z-score = -1.27, R) >droSec1.super_4 4803904 108 + 6179234 AUGAUGUCCAUGGUGCUGACCCGCCGCUGCUGACUG---GGCUGUGAGUGGGUCUGCAGGUGGUGCGUUACCAGAUGCUGGUGCUGCGCCGAGGCCUCACCACCGAUAAUC ....((((..((((((.((((((((((.(((.....---))).))).))))))).))((((((((((.((((((...)))))).))))))...)))).))))..))))... ( -49.30, z-score = -2.37, R) >droSim1.chrX 4154923 108 - 17042790 AUGAUGUCCAUGGUGCUGACCCGCCGCUGCUGACUG---GGCUGUGAGUGGGUCUGCAGGUGGUGCGUUACCAGAUGCUGGUGCUGCGCCGAGGCCUCACCACCGAUGAUC ..((((((..((((((.((((((((((.(((.....---))).))).))))))).))((((((((((.((((((...)))))).))))))...)))).))))..))).))) ( -48.40, z-score = -1.92, R) >anoGam1.chrX 12328900 106 + 22145176 --GACGCCCUGGUUGCCCACCAGCAGCAGAUGCUCGCCGAGCAGAAAGU---CCUGCAGCAGCCGCUCCAUCAGCCGCAGGUGCUGCGGCACGUCGUAGAACACAAUGUCC --((((....((((((......(((((....))).))...((((.....---.)))).)))))).........(((((((...))))))).))))................ ( -37.90, z-score = -0.76, R) >consensus AUGAUGUCCAUGGUGCUGACCCGCCGCUGCUGACUG___GGCUGUGAGUGGGUCUGCAGGUGGUGCGUUACCAGAUGCUGGUGCUGCGCCGAGGCCUCACCACCGAUGAUC ....((((..((((((.((((((((((................))).))))))).))((((((((((.((((((...)))))).))))))...)))).))))..))))... (-34.73 = -34.82 + 0.09)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:17:55 2011