| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,381,381 – 5,381,472 |
| Length | 91 |
| Max. P | 0.887543 |

| Location | 5,381,381 – 5,381,472 |
|---|---|
| Length | 91 |
| Sequences | 14 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 71.71 |
| Shannon entropy | 0.60388 |
| G+C content | 0.48216 |
| Mean single sequence MFE | -21.61 |
| Consensus MFE | -12.28 |
| Energy contribution | -12.11 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.887543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
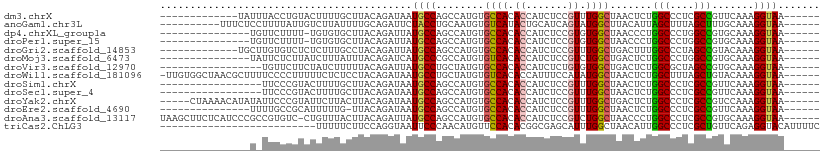
>dm3.chrX 5381381 91 + 22422827 -------------UAUUUACCUGUACUUUUGCUUACAGAUAAUGCCAGCCAUGUGCCACACCAUCUCCGUUUGGCUAACUCUGGCCCUCGCCGUUCAAAGGUAA------ -------------..........((((((((...((.(.....(((((......((((.((.......)).)))).....))))).....).)).)))))))).------ ( -21.10, z-score = -1.53, R) >anoGam1.chr3L 41273873 94 + 41284009 ----------UUUCUCCUUUUAUUGUCUUAUUUUGCAGAUUCUACCUGCAAUGUGUCAUACUGCAUCAGUAUGGCUUACAUUAGCUUUAGCUUUGCAAAGGUAA------ ----------....................(((((((((..(((...((((((((((((((((...)))))))))..))))).))..))).)))))))))....------ ( -24.50, z-score = -2.87, R) >dp4.chrXL_group1a 535974 88 - 9151740 ---------------UGUUCUUUU-UGUGUGCUUACAGAUUAUGCCAGCCAUGUGCCACACCAUCUCCGUGUGGCUAACCCUGGCCCUGGCCGUGCAAAGGUAA------ ---------------.....((((-((..((....(((.....(((((......(((((((.......))))))).....))))).)))..))..))))))...------ ( -27.20, z-score = -1.15, R) >droPer1.super_15 1169355 88 - 2181545 ---------------UGUUCUUUU-UGUGUGCUUACAGAUUAUGCCAGCCAUGUGCCACACCAUCUCCGUGUGGCUAACCCUGGCCCUGGCCGUGCAAAGGUAA------ ---------------.....((((-((..((....(((.....(((((......(((((((.......))))))).....))))).)))..))..))))))...------ ( -27.20, z-score = -1.15, R) >droGri2.scaffold_14853 6183017 91 + 10151454 -------------UGCUUGUGUCUCUCUUUGCCUACAGAUUAUGCCAGCCAUGUGCCACACCAUCUCCGUUUGGCUGACUUUGGCCCUAGCCGUACAAAGGUAA------ -------------...............((((((...........((((((..((............))..)))))).....(((....)))......))))))------ ( -17.90, z-score = 1.08, R) >droMoj3.scaffold_6473 10852054 89 + 16943266 ---------------UAUUCUCUUAUCUUUAUUUACAGAUCAUGCCCGCCAUGUGUCACACCAUCUCCGUCUGGCUGACUCUGGCCCUGGCCGUGCAAAGGUAA------ ---------------........(((((((..........((((((.((((.(.((((..(((........))).)))).)))))...)).)))).))))))).------ ( -18.60, z-score = -0.19, R) >droVir3.scaffold_12970 1328119 87 - 11907090 -----------------UGUUCUUCUAUCUUUUUACAGAUUAUGCCUGCUAUGUGCCACACCAUCUCUGUGUGGCUGACUCUGGCGCUAGCCGUGCAAAGGUAA------ -----------------........(((((((.(((.(.(((((((........(((((((.......))))))).......)))).)))).))).))))))).------ ( -24.06, z-score = -1.41, R) >droWil1.scaffold_181096 10097559 103 + 12416693 -UUGUGGCUAACGCUUUUCCCCUUUUUCUCUCCUACAGAUAAUGCCUGCUAUGUGUCACACCAUUUCCAUAUGGCUAACUCUGGCUUUAGCUGUACAAAGGUAA------ -....(((....))).....(((((........(((((.(((.(((.((((((((............)))))))).......))).))).))))).)))))...------ ( -21.30, z-score = -0.96, R) >droSim1.chrX 4149073 87 + 17042790 -----------------UUCCCGUACUUUUGCUUACAGAUAAUGCCAGCCAUGUGCCACACCAUCUCCGUUUGGCUAACUCUGGCCCUCGCCGUUCAAAGGUAA------ -----------------......((((((((...((.(.....(((((......((((.((.......)).)))).....))))).....).)).)))))))).------ ( -21.10, z-score = -1.66, R) >droSec1.super_4 4799706 87 - 6179234 -----------------UUCCCGUACUUUUGCUUACAGAUAAUGCCAGCCAUGUGCCACACCAUCUCCGUUUGGCUAACUCUGGCCCUCGCCGUUCAAAGGUAA------ -----------------......((((((((...((.(.....(((((......((((.((.......)).)))).....))))).....).)).)))))))).------ ( -21.10, z-score = -1.66, R) >droYak2.chrX 18685188 99 - 21770863 -----CUAAAACAUAUAUUCCCGUAUUCUUACUUACAGAUAAUGCCAGCCAUGUGCCACACCAUCUCCGUUUGGCUGACUCUGGCCCUCGCCGUCCAAAGGUAA------ -----.................(((........))).......(((((......((((.((.......)).)))).....)))))....(((.......)))..------ ( -17.20, z-score = -0.22, R) >droEre2.scaffold_4690 2733442 88 + 18748788 ---------------UUUUGCCGCAUUUUUG-UUACAGAUAAUGCCAGCCAUGUGCCACACCAUCUCCGUUUGGCUAACUCUGGCCCUCGCCGUUCAAAGGUAA------ ---------------..((((((((((((((-...)))).)))))..((((.((((((.((.......)).))))..))..))))..............)))))------ ( -19.20, z-score = -0.36, R) >droAna3.scaffold_13117 4870293 103 - 5790199 UAAGCUUCUCAUCCCGCCGUGUC-CUGUUUACUUACAGAUUAUGCCAGCCAUGUGCCACACCAUCUCCGUCUGGCUAACCCUGGCCCUCGCCGUGCAAAGGUAA------ ...............((((.((.-((((......)))).....(((((......((((.((.......)).)))).....)))))....)))).))........------ ( -21.40, z-score = -0.06, R) >triCas2.ChLG3 24744862 84 - 32080666 --------------------------UUUUUCUUCCAGGUAAUUCCCAACAUGUUCCACACGGCGAGCAUUUGGCUAACAUUGGCCCUCGCUGUUCAGAGGUACAUUUUC --------------------------...........((......))...((((.((.(((((((((.....(((((....)))))))))))))...).)).)))).... ( -20.70, z-score = -1.81, R) >consensus _______________U_UUCCCUUAUUUUUGCUUACAGAUAAUGCCAGCCAUGUGCCACACCAUCUCCGUUUGGCUAACUCUGGCCCUCGCCGUGCAAAGGUAA______ ..........................................((((........((((.((.......)).)))).......(((....))).......))))....... (-12.28 = -12.11 + -0.18)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:17:53 2011