
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,371,231 – 5,371,360 |
| Length | 129 |
| Max. P | 0.748227 |

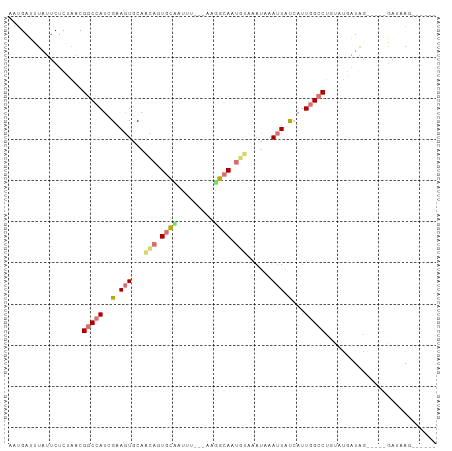
| Location | 5,371,231 – 5,371,330 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 60.03 |
| Shannon entropy | 0.62689 |
| G+C content | 0.36940 |
| Mean single sequence MFE | -20.55 |
| Consensus MFE | -9.12 |
| Energy contribution | -10.88 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.748227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5371231 99 + 22422827 AAUGAUUUAUUCUCUAACGGCCAUCGUAGUGCAGCAGUGCCAUUUUUUAAGGCAAUCUAAAUAAAUUAUCAUUGGCCUUAUUGGUAA-----GGUUAGUUAAAG .((((((((((.....(((.....))).....((...((((.........))))..)).)))))))))).((((((((((....)))-----)))))))..... ( -18.60, z-score = -0.37, R) >droEre2.scaffold_4690 2727308 101 + 18748788 AUUGAUUUCUUCUCUAACGGCCAUCGAAGUGCAGCAGUGCUAUUU---CAGGCACUGUUAAUAAAUUAUCAUUGGCCUGUAUUAUGGCCCGUAACAAGCGAAAG ..................((((((...(((((((.((((((....---..))))))((((((........)))))))))))))))))))(((.....))).... ( -26.10, z-score = -1.59, R) >droYak2.chrX 18678878 90 - 21770863 CAUGAUUUAUUCUCUAUCGGCCAUCGAAGUGCAACAGUGCGAUUU---AAAGCACUGUAAAUAAAUUAUCAUUGGCCUAUAUGAUGG-----GAAAAG------ .........(((.((((((((((..(((((...(((((((.....---...)))))))......))).))..))))).....)))))-----)))...------ ( -24.80, z-score = -3.10, R) >droMoj3.scaffold_6473 10839982 91 + 16943266 CAAGAUUUAUGCCUUCACG-CAAUAGAAAUUCAAUCGUUAAAAAUU--CAUUCAAUGCUGACCUAAUUCGCCUGGGCAGCUAAAUAU----UGUUUAG------ (((.((((((((......)-))........................--........((((.((((.......))))))))))))).)----)).....------ ( -12.70, z-score = 0.18, R) >consensus AAUGAUUUAUUCUCUAACGGCCAUCGAAGUGCAACAGUGCAAUUU___AAGGCAAUGUAAAUAAAUUAUCAUUGGCCUGUAUGAUAG_____GAUAAG______ ..................(((((..(((((...((((((((.........))))))))......)))))...)))))........................... ( -9.12 = -10.88 + 1.75)


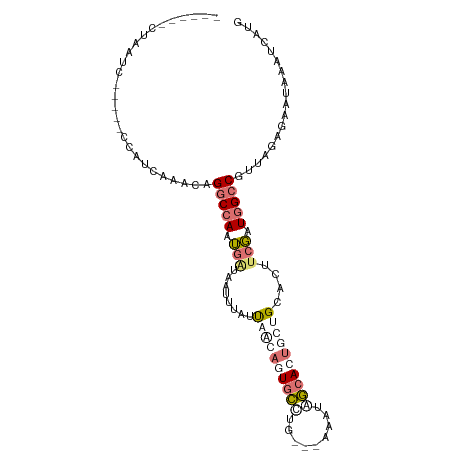
| Location | 5,371,231 – 5,371,330 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 60.03 |
| Shannon entropy | 0.62689 |
| G+C content | 0.36940 |
| Mean single sequence MFE | -20.47 |
| Consensus MFE | -7.50 |
| Energy contribution | -10.25 |
| Covariance contribution | 2.75 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.682219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5371231 99 - 22422827 CUUUAACUAACC-----UUACCAAUAAGGCCAAUGAUAAUUUAUUUAGAUUGCCUUAAAAAAUGGCACUGCUGCACUACGAUGGCCGUUAGAGAAUAAAUCAUU (((((((.....-----(((....)))(((((((((....)))).(((..((((.........))))...)))........))))))))))))........... ( -18.10, z-score = -1.50, R) >droEre2.scaffold_4690 2727308 101 - 18748788 CUUUCGCUUGUUACGGGCCAUAAUACAGGCCAAUGAUAAUUUAUUAACAGUGCCUG---AAAUAGCACUGCUGCACUUCGAUGGCCGUUAGAGAAGAAAUCAAU .((((((((((((..((((........))))..))))))........((((((...---.....))))))..)).(((..((....))..)))..))))..... ( -25.30, z-score = -1.43, R) >droYak2.chrX 18678878 90 + 21770863 ------CUUUUC-----CCAUCAUAUAGGCCAAUGAUAAUUUAUUUACAGUGCUUU---AAAUCGCACUGUUGCACUUCGAUGGCCGAUAGAGAAUAAAUCAUG ------......-----...((.(((.(((((.(((..........(((((((...---.....)))))))......))).))))).)))))............ ( -21.69, z-score = -2.86, R) >droMoj3.scaffold_6473 10839982 91 - 16943266 ------CUAAACA----AUAUUUAGCUGCCCAGGCGAAUUAGGUCAGCAUUGAAUG--AAUUUUUAACGAUUGAAUUUCUAUUG-CGUGAAGGCAUAAAUCUUG ------.....((----(.(((((..((((((.((((..(((((((((.(((((..--....))))).).))))...)))))))-).))..))))))))).))) ( -16.80, z-score = -0.27, R) >consensus ______CUAAUC_____CCAUCAAACAGGCCAAUGAUAAUUUAUUAACAGUGCCUG___AAAUAGCACUGCUGCACUUCGAUGGCCGUUAGAGAAUAAAUCAUG ...........................(((((.(((........((((((((((.........))))))))))....))).))))).................. ( -7.50 = -10.25 + 2.75)

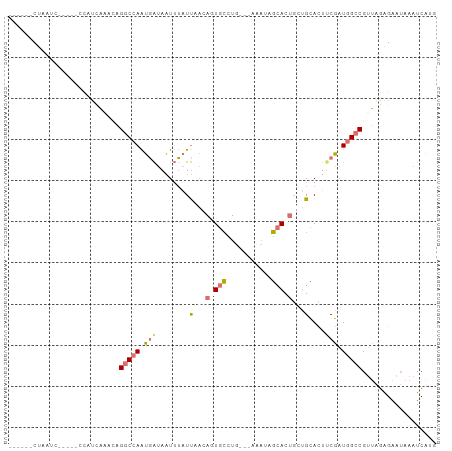
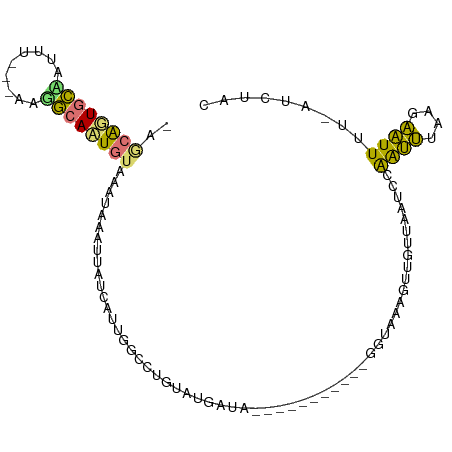
| Location | 5,371,263 – 5,371,360 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 56.88 |
| Shannon entropy | 0.68213 |
| G+C content | 0.32790 |
| Mean single sequence MFE | -21.38 |
| Consensus MFE | -5.49 |
| Energy contribution | -4.30 |
| Covariance contribution | -1.19 |
| Combinations/Pair | 1.83 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.596566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5371263 97 + 22422827 -AGCAGUGCCAUUUUUUAAGGCAAUCUAAAUAAAUUAUCAUUGGCCUUAUUGGUAA-----GGUUAGUUAAAGUUGUUAAUCGGAUUUAAGAAUUUU-AUCUAU -.((..(((((.....((((((.....................)))))).))))).-----.))(((.(((((((.((((......)))).))))))-).))). ( -20.50, z-score = -1.90, R) >droEre2.scaffold_4690 2727340 100 + 18748788 -AGCAGUGCUAUUU---CAGGCACUGUUAAUAAAUUAUCAUUGGCCUGUAUUAUGGCCCGUAACAAGCGAAAGUUGUUAAUCCAAUUUCCGAAUUUUUAUCUAC -(((((((((....---..))))))))).(((((...((...((((........))))..((((((.(....)))))))...........))...))))).... ( -26.70, z-score = -3.42, R) >droYak2.chrX 18678910 88 - 21770863 -AACAGUGCGAUUU---AAAGCACUGUAAAUAAAUUAUCAUUGGCCUAUAUGAUG-----------GGAAAAGUUGUUAAUCCAAUUUUGGAAUUUU-AUCAAC -.(((((((.....---...))))))).......(((((((........))))))-----------).....((((.(((((((....))))...))-).)))) ( -20.10, z-score = -2.45, R) >droMoj3.scaffold_6473 10840013 90 + 16943266 AAUCGUUAAAAAUU---CAUUCAAUGCUGACCUAAUUCGCCUGGGCAGCUAAAUAU----------UGUUUAGUGGCCAAUGCGGCGUAUGCGUUAU-AUUUAU ..............---.....(((((..........((((..(((.(((((((..----------.))))))).))).....))))...)))))..-...... ( -18.22, z-score = 0.22, R) >consensus _AGCAGUGCAAUUU___AAGGCAAUGUAAAUAAAUUAUCAUUGGCCUGUAUGAUA___________GGUAAAGUUGUUAAUCCAAUUUAAGAAUUUU_AUCUAC ..((((((((.........))))))))........................................................((((....))))......... ( -5.49 = -4.30 + -1.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:17:52 2011