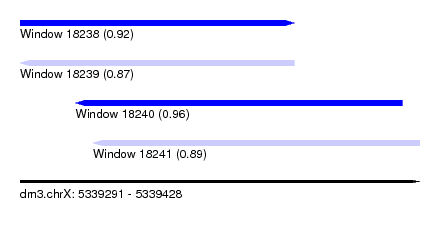
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,339,291 – 5,339,428 |
| Length | 137 |
| Max. P | 0.955770 |

| Location | 5,339,291 – 5,339,385 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 72.44 |
| Shannon entropy | 0.47672 |
| G+C content | 0.45941 |
| Mean single sequence MFE | -20.82 |
| Consensus MFE | -12.86 |
| Energy contribution | -12.66 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.923077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
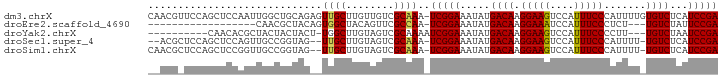
>dm3.chrX 5339291 94 + 22422827 CAACGUUCCAGCUCCAAUUGGCUGCAGAGUUGCUUGUUGUCGCAAA-UCGGAAAUAUGACAAGGAAGUCCAUUUCCCAUUUUGUGUCUCAUCCGA ((((....(((((......)))))....))))......(.((((((-..((((((..(((......))).))))))...)))))).)........ ( -22.40, z-score = -0.69, R) >droEre2.scaffold_4690 2694808 73 + 18748788 ------------------CAACGCUACAGUGGCUACAGUUCGCCAA-UCGGAAAUAUGACAAGGAAAUCCAUUUCCCUCU---UGUCUAUUCCGA ------------------...........((((........)))).-((((((....((((((((((....)))))...)---))))..)))))) ( -19.40, z-score = -2.73, R) >droYak2.chrX 18645908 81 - 21770863 ----------CAACACGCUACUACUACU-UGGCUUGUAGUCGCAAAAUCGGAAAUAUGACAAGGAAGUCCAUUUCCCCUU---UGUCUAAUCCGA ----------......((.(((((....-......))))).))....(((((.....((((((((((....)))))...)---))))...))))) ( -20.40, z-score = -2.77, R) >droSec1.super_4 4762998 89 - 6179234 --ACGCUCCAGCUCCAGUUGCCGGUAG--UUGCUUGUAGUCGCAAA-UCGGAAAUAUGACAAGGAAGUCCAUUUCCCAUUUU-UGUCUCAUCCGA --..(((((.((.......)).)).))--)(((........)))..-(((((.....((((((((((....))))).....)-))))...))))) ( -19.30, z-score = -0.55, R) >droSim1.chrX 4111203 91 + 17042790 CAACGCUCCAGCUCCGGUUGCCGGUAG--UUGCUUGUAGUCGCAAA-UCGGAAAUAUGACAAGGAAGUCCAUUUCCCAUUUU-UGUCUCAUCCGA ....((..(((((((((...)))).))--)))...)).........-(((((.....((((((((((....))))).....)-))))...))))) ( -22.60, z-score = -1.09, R) >consensus __ACG_UCCAGCUCCAGUUGCCGGUAG__UUGCUUGUAGUCGCAAA_UCGGAAAUAUGACAAGGAAGUCCAUUUCCCAUUUU_UGUCUCAUCCGA .............................((((........))))..(((((.....((((.(((((....))))).......))))...))))) (-12.86 = -12.66 + -0.20)
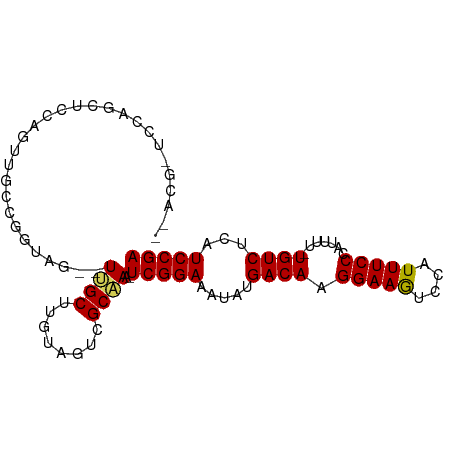
| Location | 5,339,291 – 5,339,385 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 72.44 |
| Shannon entropy | 0.47672 |
| G+C content | 0.45941 |
| Mean single sequence MFE | -22.42 |
| Consensus MFE | -14.56 |
| Energy contribution | -14.52 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.872489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5339291 94 - 22422827 UCGGAUGAGACACAAAAUGGGAAAUGGACUUCCUUGUCAUAUUUCCGA-UUUGCGACAACAAGCAACUCUGCAGCCAAUUGGAGCUGGAACGUUG .((...(((.....((((.((((((((((......))).))))))).)-)))((........))..)))..((((........))))...))... ( -22.70, z-score = -0.72, R) >droEre2.scaffold_4690 2694808 73 - 18748788 UCGGAAUAGACA---AGAGGGAAAUGGAUUUCCUUGUCAUAUUUCCGA-UUGGCGAACUGUAGCCACUGUAGCGUUG------------------ ((((((..((((---((.(..(......)..)))))))....))))))-.((((........))))...........------------------ ( -21.70, z-score = -2.09, R) >droYak2.chrX 18645908 81 + 21770863 UCGGAUUAGACA---AAGGGGAAAUGGACUUCCUUGUCAUAUUUCCGAUUUUGCGACUACAAGCCA-AGUAGUAGUAGCGUGUUG---------- (((((.((((((---...(((((......))))))))).))..)))))....((.(((((......-....))))).))......---------- ( -20.60, z-score = -1.32, R) >droSec1.super_4 4762998 89 + 6179234 UCGGAUGAGACA-AAAAUGGGAAAUGGACUUCCUUGUCAUAUUUCCGA-UUUGCGACUACAAGCAA--CUACCGGCAACUGGAGCUGGAGCGU-- (((((...((((-.....(((((......))))))))).....)))))-.((((........))))--...(((((.......))))).....-- ( -23.40, z-score = -1.35, R) >droSim1.chrX 4111203 91 - 17042790 UCGGAUGAGACA-AAAAUGGGAAAUGGACUUCCUUGUCAUAUUUCCGA-UUUGCGACUACAAGCAA--CUACCGGCAACCGGAGCUGGAGCGUUG ...((((((...-.((((.((((((((((......))).))))))).)-)))((........))..--)).(((((.......)))))..)))). ( -23.70, z-score = -1.14, R) >consensus UCGGAUGAGACA_AAAAUGGGAAAUGGACUUCCUUGUCAUAUUUCCGA_UUUGCGACUACAAGCAA__CUACCGGCAACUGGAGCUGGA_CGU__ (((((...((((......(((((......))))))))).....)))))..((((........))))............................. (-14.56 = -14.52 + -0.04)

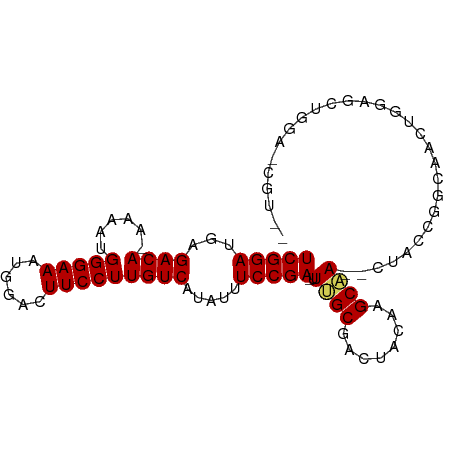
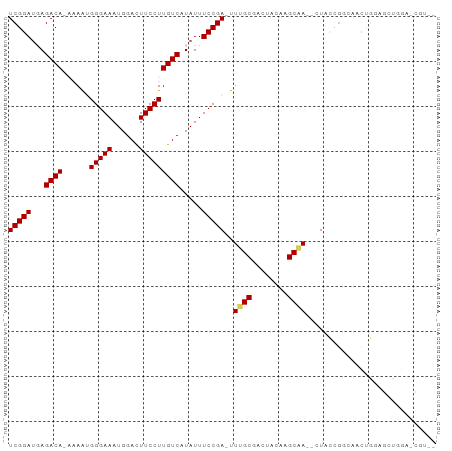
| Location | 5,339,310 – 5,339,422 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 126 |
| Reading direction | reverse |
| Mean pairwise identity | 67.21 |
| Shannon entropy | 0.50802 |
| G+C content | 0.46668 |
| Mean single sequence MFE | -29.22 |
| Consensus MFE | -16.94 |
| Energy contribution | -16.74 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.955770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
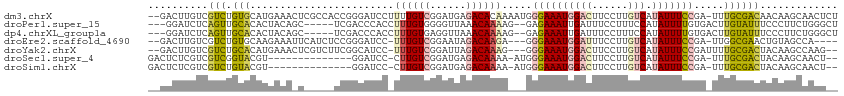
>dm3.chrX 5339310 112 - 22422827 UCGUCUGUGCAUGAAACUCGCCACCGGGAUCC-------------UUUUGUCGGAUGAGACACAAAAUGGGAAAUGGACUUCCUUGUCAUAUUUCCGA-UUUGCGACAACAAGCAACUCUGCAGCC ..((((((((......((((....))))((((-------------.......))))..).))))((((.((((((((((......))).))))))).)-)))..))).....(((....))).... ( -28.60, z-score = -0.47, R) >droEre2.scaffold_4690 2694819 98 - 18748788 --------------UCGUCUGUGCAAGAAAUUCAUCUCCGGGAUCCUUUGUCGGAAUAGACA---AGAGGGAAAUGGAUUUCCUUGUCAUAUUUCCGA-UUGGCGAACUGUAGCCA---------- --------------(((..((((((((((((((((.(((((...))((((((......))))---))..))).)))))))).)))).))))....)))-.((((........))))---------- ( -28.60, z-score = -1.02, R) >droYak2.chrX 18645924 101 + 21770863 --------------UCGUCUGCACAUGAAACUCGUCUUCGGCAUCCUUUGUCGGAUUAGACA---AAGGGGAAAUGGACUUCCUUGUCAUAUUUCCGAUUUUGCGACUACAAGCCAAG-------- --------------..(((.(((..........(.(....))..((((((((......))))---))))((((((((((......))).))))))).....))))))...........-------- ( -29.10, z-score = -2.16, R) >droSec1.super_4 4763015 94 + 6179234 --------------UCGUCGGUACGUGGAUCC--------------CUUGUCGGAUGAGACA-AAAAUGGGAAAUGGACUUCCUUGUCAUAUUUCCGA-UUUGCGACUACAAGCAACUAC-CGGC- --------------..(((((((((..((((.--------------.(((((......))))-).....((((((((((......))).)))))))))-))..))............)))-))))- ( -31.10, z-score = -2.63, R) >droSim1.chrX 4111222 94 - 17042790 --------------UCGUCUGUACGUGGAUCC--------------CUUGUCGGAUGAGACA-AAAAUGGGAAAUGGACUUCCUUGUCAUAUUUCCGA-UUUGCGACUACAAGCAACUAC-CGGC- --------------..(..((((((..((((.--------------.(((((......))))-).....((((((((((......))).)))))))))-))..))..))))..)......-....- ( -28.70, z-score = -2.30, R) >consensus ______________UCGUCUGUACAUGGAUCC______________UUUGUCGGAUGAGACA_AAAAUGGGAAAUGGACUUCCUUGUCAUAUUUCCGA_UUUGCGACUACAAGCAACU___C_GC_ ................................................((((......)))).......((((((((((......))).)))))))....((((........)))).......... (-16.94 = -16.74 + -0.20)
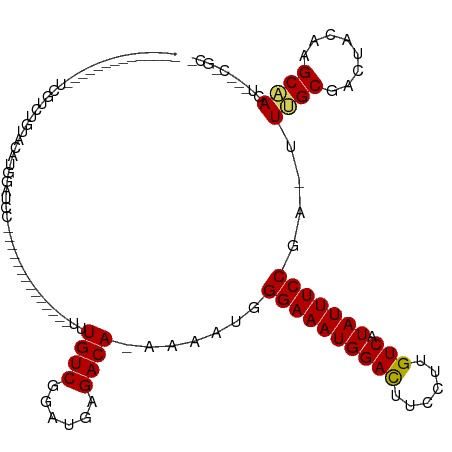
| Location | 5,339,316 – 5,339,428 |
|---|---|
| Length | 112 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 62.78 |
| Shannon entropy | 0.68225 |
| G+C content | 0.44984 |
| Mean single sequence MFE | -29.11 |
| Consensus MFE | -11.17 |
| Energy contribution | -11.26 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.892143 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
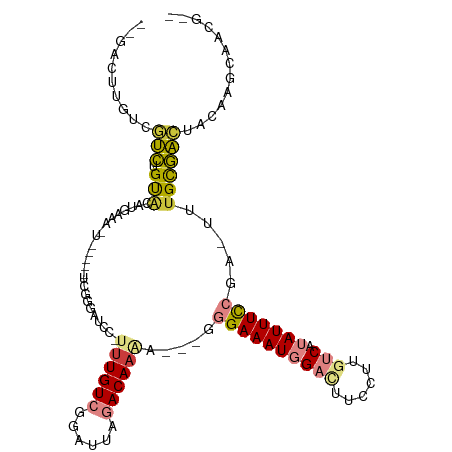
>dm3.chrX 5339316 112 - 22422827 --GACUUGUCGUCUGUGCAUGAAACUCGCCACCGGGAUCCUUUUGUCGGAUGAGACACAAAAUGGGAAAUGGACUUCCUUGUCAUAUUUCCGA-UUUGCGACAACAAGCAACUCU --(.(((((.((((((((......((((....))))((((.......))))..).))))((((.((((((((((......))).))))))).)-)))..))).))))))...... ( -31.80, z-score = -1.71, R) >droPer1.super_15 1124741 105 + 2181545 ---GGAUCUCAGUUGCACACUACAGC-----UCGACCCACCUUUGUGGGGUUAAACAAAAG--GAGAAAUUGAUUUCCUUUCCAUAUUUUGUGACUUGUAUUUCCCUUCUGGGCU ---....(((((........(((((.-----(((.(((((....)))))........((((--((((......))))))))..........))).)))))........))))).. ( -23.69, z-score = -0.01, R) >dp4.chrXL_group1a 490482 105 + 9151740 ---GGAUCUCAGUUGCACACUACAGC-----UCGACCCACCUUUGUGAGGUUAAACAAAAG--GAGAAAUUGAUUUCCUUUCCAUAUUUUGUGACUUGUAUUUCCCUUCUGGGCU ---.......(((..((.........-----..(((((((....))).)))).....((((--((((......))))))))........))..))).......(((....))).. ( -21.90, z-score = 0.06, R) >droEre2.scaffold_4690 2694819 104 - 18748788 --GACUUGUCGUCUGUGCAAGAAAUUCAUCUCCGGGAUCC-UUUGUCGGAAUAGACAAGA---GGGAAAUGGAUUUCCUUGUCAUAUUUCCGA-UUGGCGAACUGUAGCCA---- --..((((((.((((.(((((..((((.......))))..-)))))))))...)))))).---.((((((((((......))).)))))))..-.((((........))))---- ( -30.90, z-score = -1.18, R) >droYak2.chrX 18645924 107 + 21770863 --GACUUGUCGUCUGCACAUGAAACUCGUCUUCGGCAUCC-UUUGUCGGAUUAGACAAAG---GGGAAAUGGACUUCCUUGUCAUAUUUCCGAUUUUGCGACUACAAGCCAAG-- --(.(((((.(((.(((..........(.(....))..((-((((((......)))))))---)((((((((((......))).))))))).....)))))).))))).)...-- ( -35.10, z-score = -3.35, R) >droSec1.super_4 4763021 96 + 6179234 GACUCUCGUCGUCGGUACGU--------------GGAUCC-CUUGUCGGAUGAGACAAAA-AUGGGAAAUGGACUUCCUUGUCAUAUUUCCGA-UUUGCGACUACAAGCAACU-- (((....)))((.(((.((.--------------.((((.-.(((((......)))))..-...((((((((((......))).)))))))))-))..)))))))........-- ( -29.60, z-score = -2.04, R) >droSim1.chrX 4111228 96 - 17042790 GACUCUCGUCGUCUGUACGU--------------GGAUCC-CUUGUCGGAUGAGACAAAA-AUGGGAAAUGGACUUCCUUGUCAUAUUUCCGA-UUUGCGACUACAAGCAACU-- (((....)))(..((((((.--------------.((((.-.(((((......)))))..-...((((((((((......))).)))))))))-))..))..))))..)....-- ( -30.80, z-score = -2.86, R) >consensus __GACUUGUCGUCUGUACAUGAAA_U_____UCGGGAUCC_UUUGUCGGAUUAGACAAAA___GGGAAAUGGACUUCCUUGUCAUAUUUCCGA_UUUGCGACUACAAGCAACG__ ..........(((.(((........................((((((......)))))).....((((((((((......))).))))))).....))))))............. (-11.17 = -11.26 + 0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:17:48 2011