
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,309,116 – 5,309,176 |
| Length | 60 |
| Max. P | 0.999858 |

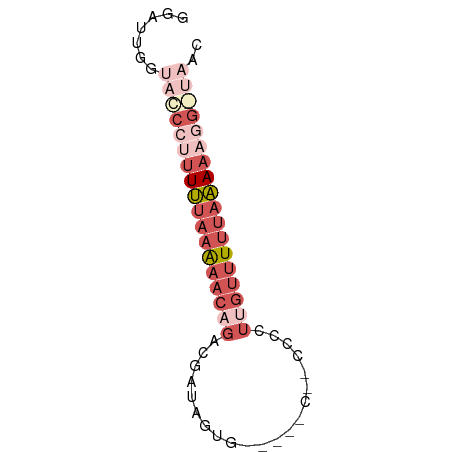
| Location | 5,309,116 – 5,309,176 |
|---|---|
| Length | 60 |
| Sequences | 5 |
| Columns | 66 |
| Reading direction | forward |
| Mean pairwise identity | 58.49 |
| Shannon entropy | 0.72167 |
| G+C content | 0.36627 |
| Mean single sequence MFE | -17.16 |
| Consensus MFE | -4.61 |
| Energy contribution | -7.57 |
| Covariance contribution | 2.96 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.983547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
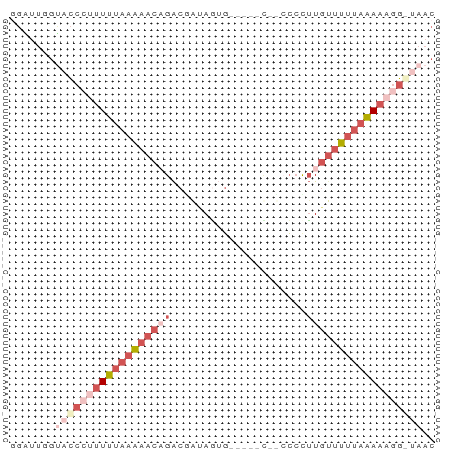
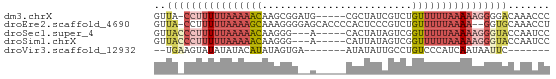
>dm3.chrX 5309116 60 + 22422827 GGGUUUGUCCCCUUUUUAAAAACAGACGAUAGCG-----CAUCCGCUUGUUUUUAAAAAGG-UAAC ......((..((((((((((((((......((((-----....))))))))))))))))))-..)) ( -17.60, z-score = -3.03, R) >droEre2.scaffold_4690 2663407 63 + 18748788 AGGUUUGCACC--UUUUAAAAACAGACGGGAGUGGGGUGCUCCCCUUUGCUUUUAAAAAGG-UAAC ........(((--((((.((((((((.((((((.....)))))).)))).)))).))))))-)... ( -21.80, z-score = -2.19, R) >droSec1.super_4 4732789 58 - 6179234 GGAUUGGUACCCUUUUUAAAAACCGACUAUAGUG-----U---CCCUUGUUUUUAAAAAGGGUAAC .......((((((((((((((((.(((......)-----)---)....)))))))))))))))).. ( -21.30, z-score = -4.28, R) >droSim1.chrX 4085632 58 + 17042790 GGAUUGGUACCCUUUUUAAAAACCGACUAUAAUG-----U---CCCUUGUUUUUAAAAAGGGUAAC .......((((((((((((((((.(((......)-----)---)....)))))))))))))))).. ( -21.30, z-score = -4.68, R) >droVir3.scaffold_12932 1853515 50 - 2102469 -------GAAUUAUUGAUGGGACAGGCAAUAUAU-------UCACUAUAUGUAUAUAUACUUCA-- -------(((((((((.((...))..))))).))-------)).....................-- ( -3.80, z-score = 0.87, R) >consensus GGAUUGGUACCCUUUUUAAAAACAGACGAUAGUG_____C__CCCCUUGUUUUUAAAAAGG_UAAC .......((((((((((((((((.........................)))))))))))))))).. ( -4.61 = -7.57 + 2.96)

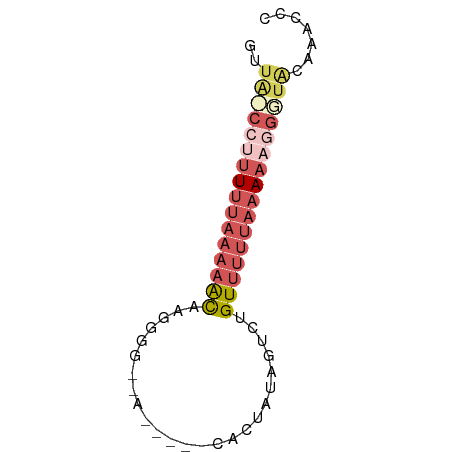
| Location | 5,309,116 – 5,309,176 |
|---|---|
| Length | 60 |
| Sequences | 5 |
| Columns | 66 |
| Reading direction | reverse |
| Mean pairwise identity | 58.49 |
| Shannon entropy | 0.72167 |
| G+C content | 0.36627 |
| Mean single sequence MFE | -18.74 |
| Consensus MFE | -6.55 |
| Energy contribution | -9.03 |
| Covariance contribution | 2.48 |
| Combinations/Pair | 1.31 |
| Mean z-score | -3.78 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.60 |
| SVM RNA-class probability | 0.999858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5309116 60 - 22422827 GUUA-CCUUUUUAAAAACAAGCGGAUG-----CGCUAUCGUCUGUUUUUAAAAAGGGGACAAACCC ((..-((((((((((((((..(((...-----.....)))..))))))))))))))..))...... ( -22.30, z-score = -4.87, R) >droEre2.scaffold_4690 2663407 63 - 18748788 GUUA-CCUUUUUAAAAGCAAAGGGGAGCACCCCACUCCCGUCUGUUUUUAAAA--GGUGCAAACCU ((.(-((((((.(((((((.(.(((((.......))))).).)))))))))))--)))))...... ( -22.70, z-score = -3.87, R) >droSec1.super_4 4732789 58 + 6179234 GUUACCCUUUUUAAAAACAAGGG---A-----CACUAUAGUCGGUUUUUAAAAAGGGUACCAAUCC (.((((((((((((((((....(---(-----(......))).)))))))))))))))).)..... ( -22.00, z-score = -4.97, R) >droSim1.chrX 4085632 58 - 17042790 GUUACCCUUUUUAAAAACAAGGG---A-----CAUUAUAGUCGGUUUUUAAAAAGGGUACCAAUCC (.((((((((((((((((....(---(-----(......))).)))))))))))))))).)..... ( -22.00, z-score = -5.29, R) >droVir3.scaffold_12932 1853515 50 + 2102469 --UGAAGUAUAUAUACAUAUAGUGA-------AUAUAUUGCCUGUCCCAUCAAUAAUUC------- --...(((((((.(((.....))).-------)))))))....................------- ( -4.70, z-score = 0.08, R) >consensus GUUA_CCUUUUUAAAAACAAGGGG__A_____CACUAUAGUCUGUUUUUAAAAAGGGUACAAACCC ..((((((((((((((((.........................))))))))))))))))....... ( -6.55 = -9.03 + 2.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:17:42 2011