
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,297,004 – 5,297,097 |
| Length | 93 |
| Max. P | 0.594834 |

| Location | 5,297,004 – 5,297,097 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 76.77 |
| Shannon entropy | 0.42411 |
| G+C content | 0.31776 |
| Mean single sequence MFE | -10.63 |
| Consensus MFE | -9.37 |
| Energy contribution | -9.48 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.50 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.594834 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
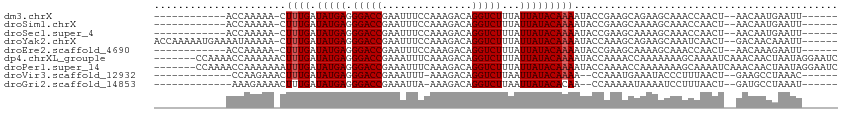
>dm3.chrX 5297004 93 + 22422827 ------------ACCAAAAA-CUUUGAUAUGAGGGACCGAAUUUCCAAAGACAGGUCUUUAUUAUACAAAAUACCGAAGCAGAAGCAAACCAACU--AACAAUGAAUU------ ------------........-.((((.(((((.(((((...............)))))...)))))))))........((....)).........--...........------ ( -10.86, z-score = -0.51, R) >droSim1.chrX 4073884 93 + 17042790 ------------ACCAAAAA-CUUUGAUAUGAGGGACCGAAUUUCCAAAGACAGGUCUUUAUUAUACAAAAUACCGAAGCAAAAGCAAACCAACU--AACAAUGAAUU------ ------------........-.((((.(((((.(((((...............)))))...)))))))))........((....)).........--...........------ ( -10.86, z-score = -0.63, R) >droSec1.super_4 4721083 93 - 6179234 ------------ACCAAAAA-CUUUGAUAUGAGGGACCGAAUUUCCAAAGACAGGUCUUUAUUAUACAAAAUACCGAAGCAAAAGCAAACCAACU--AACAAUGAAUU------ ------------........-.((((.(((((.(((((...............)))))...)))))))))........((....)).........--...........------ ( -10.86, z-score = -0.63, R) >droYak2.chrX 18598112 105 - 21770863 ACCAAAAAUGAAAAUAAAAA-CUUUGAUAUGAGGGACCGAAUUUCCAAAGACAGGUCUUUAUUAUACAAAAUACCAAAGCAGAAGCAAAUCAACU--GACAACAAAUU------ ........(((.........-.((((.(((((.(((((...............)))))...)))))))))........((....))...)))...--...........------ ( -11.96, z-score = -0.65, R) >droEre2.scaffold_4690 2651088 93 + 18748788 ------------ACCAAAAA-CUUUGAUAUGAGGGACCGAAUUUCCAAAGACAGGUCUUUAUUAUACAAAAUACCGAAGCAAAAGCAAACCAACU--AACAAAGAAUU------ ------------........-(((((.(((((.(((((...............)))))...)))))............((....)).........--..)))))....------ ( -13.06, z-score = -1.73, R) >dp4.chrXL_group1e 10889011 107 + 12523060 -------CCAAAACCAAAAAACUUUGAUAUGAGGGACCGAAAUUUCAAAGACAGGUCUUUAUUAUACAAAAUACCAAAACCAAAAAAAGCAAAAUCAAACAACUAAUAGGAAUC -------......((......((((((.((..(....)...)).))))))...(((.(((........))).))).................................)).... ( -10.70, z-score = -0.48, R) >droPer1.super_14 330992 107 + 2168203 -------CCAAAACCAAAAAAAUUUGAUAUGAGGGACCGAAAUUUCAAAGACAGGUCUUUAUUAUACAAAAUACCAAAACCAAAAAAAGCAAAAUCAAACAACUAAUAGGAAUC -------...............((((.(((((.(((((...............)))))...)))))))))............................................ ( -9.86, z-score = -0.17, R) >droVir3.scaffold_12932 1835223 90 - 2102469 -------------CCAAGAAACUUUGAUAUGAGGGACCGAAAUUU-AAAGACAGGUCUUAAUUAUACAAAA--CCAAAUGAAAUACCCUUUAACU--GAAGCCUAAAC------ -------------.........((((.(((((.(((((.......-.......)))))...))))))))).--......................--...........------ ( -9.74, z-score = -0.18, R) >droGri2.scaffold_14853 2512248 90 + 10151454 -------------AAAGAAAACUUUGAUAUGAGGGACCGAAAUUA-AAAGACAGGUCUUAAUUAUACACAA--CCAAAAAUAAAAUCCUUUAACU--GAUGCCUAAAU------ -------------...........((.(((((.(((((.......-.......)))))...)))))))...--......................--...........------ ( -7.74, z-score = 0.44, R) >consensus ____________ACCAAAAA_CUUUGAUAUGAGGGACCGAAUUUCCAAAGACAGGUCUUUAUUAUACAAAAUACCAAAGCAAAAGCAAACCAACU__AACAACGAAUU______ ......................((((.(((((.(((((...............)))))...)))))))))............................................ ( -9.37 = -9.48 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:17:38 2011