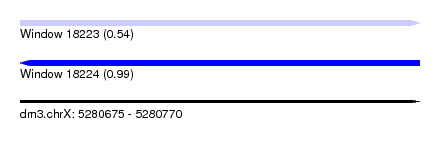
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,280,675 – 5,280,770 |
| Length | 95 |
| Max. P | 0.988777 |

| Location | 5,280,675 – 5,280,770 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 62.55 |
| Shannon entropy | 0.64227 |
| G+C content | 0.43746 |
| Mean single sequence MFE | -28.05 |
| Consensus MFE | -5.89 |
| Energy contribution | -7.45 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.21 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.538145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
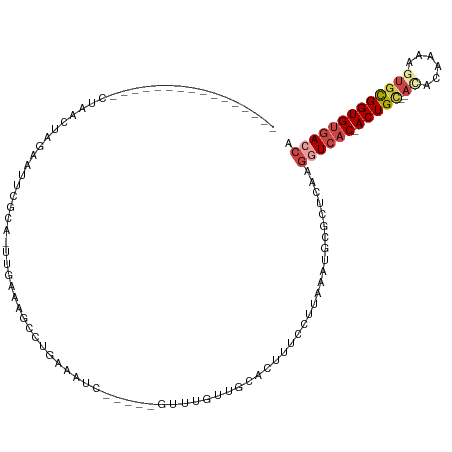
>dm3.chrX 5280675 95 + 22422827 UGGUCACACCGCACUUUUGUGU--ACAGU-GUGACCUUGAGCGCAUUUAAGGAAAGUGCAAGAAAC-----GAUUUCAGGCUUCCAA-UGCCAAUUAUAGUUAG---------------- .((((((((.((((....))))--...))-))))))...(((((((((.....)))))).......-----.......(((......-.))).......)))..---------------- ( -28.50, z-score = -2.58, R) >droEre2.scaffold_4690 2638044 106 + 18748788 UGGUCACACCGCACAAGAGU----GCAGU-GUGACCUUGAGCACAUCUAAAGAAAGUUCAACAAA------GUUUUCAGGUUUUCAA-UGCGUACUCCAAAUAGCCAAUUACAG--UUAG .((((((((.((((....))----)).))-))))))((((((...((....))..))))))....------.......((((.....-..............))))........--.... ( -26.91, z-score = -2.42, R) >droYak2.chrX 18584896 113 - 21770863 UGGUCACACCGCAGUCUUGUUUUUGCAGU-GUGAUCUUGAGCGCAUCUACAGAAAGUUCAACAAA------GUUUUCACGUUUUCAAAUGCGUGCGCCAAAUAGCCAAUUAUAUAUUUAG .((((((((.((((........)))).))-))))))(((.((((.......((((..(......)------..))))((((........))))))))))).................... ( -29.30, z-score = -2.39, R) >droSec1.super_4 4708197 95 - 6179234 UGGUCACACCGCACUUUUCUGU--GCAGU-GUGACCUUGAGCGCUUUUAAGGAAAGAGCAAAAAAC-----GAUUUCUGGCUUCCAA-UGCCAAUUAUAGUUAG---------------- .((((((((.((((......))--)).))-))))))...(((((((((.....)))))).......-----......((((......-.))))......)))..---------------- ( -33.10, z-score = -4.00, R) >droSim1.chrX_random 1822129 95 + 5698898 UGGUCACACCACACUUUUGUGU--GCAGU-GUGACCUUGAGCGCAUUUAAAGAAAGAGCAAAAAAC-----GAUUUCAGGCUUCCAA-UGCCAAUUAUAGUUAG---------------- .(((((((((((((....))))--)..))-))))))...(((.........((((..(.......)-----..)))).(((......-.))).......)))..---------------- ( -27.10, z-score = -2.70, R) >dp4.chrXL_group1e 10873530 106 + 12523060 CCUUAGCACCAUACGCAUGCAU--CCAGUCGGAAUCCAGCGCGCGAGUCAGCAUAGGGCAGCCAAAUGGCAAAAUCCAGGCAUUCGAAAAGGAUCCCACAGCAGUAAG------------ (((..((...((.(((.(((.(--((....))).....))).))).))..))..)))((.(((....)))........((.((((.....)))).))...))......------------ ( -23.40, z-score = 0.23, R) >consensus UGGUCACACCGCACUUUUGUGU__GCAGU_GUGACCUUGAGCGCAUUUAAAGAAAGUGCAACAAAC_____GAUUUCAGGCUUCCAA_UGCCAACUAUAAUUAG________________ .((((((((.((............)).)).))))))..........................................(((........)))............................ ( -5.89 = -7.45 + 1.56)


| Location | 5,280,675 – 5,280,770 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 62.55 |
| Shannon entropy | 0.64227 |
| G+C content | 0.43746 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -9.65 |
| Energy contribution | -10.32 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.988777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5280675 95 - 22422827 ----------------CUAACUAUAAUUGGCA-UUGGAAGCCUGAAAUC-----GUUUCUUGCACUUUCCUUAAAUGCGCUCAAGGUCAC-ACUGU--ACACAAAAGUGCGGUGUGACCA ----------------.............(((-((((((((..(((...-----..)))..))...))))...)))))......((((((-(((((--((......))))))))))))). ( -33.00, z-score = -4.38, R) >droEre2.scaffold_4690 2638044 106 - 18748788 CUAA--CUGUAAUUGGCUAUUUGGAGUACGCA-UUGAAAACCUGAAAAC------UUUGUUGAACUUUCUUUAGAUGUGCUCAAGGUCAC-ACUGC----ACUCUUGUGCGGUGUGACCA ....--......((((((((((((((....((-..(((...........------)))..))......))))))))).)).)))((((((-(((((----((....))))))))))))). ( -34.30, z-score = -3.84, R) >droYak2.chrX 18584896 113 + 21770863 CUAAAUAUAUAAUUGGCUAUUUGGCGCACGCAUUUGAAAACGUGAAAAC------UUUGUUGAACUUUCUGUAGAUGCGCUCAAGAUCAC-ACUGCAAAAACAAGACUGCGGUGUGACCA .............(((...(((((((((((((((..(.((.((....))------.)).)..)).....))).).))))).)))).((((-((((((..........))))))))))))) ( -27.70, z-score = -1.61, R) >droSec1.super_4 4708197 95 + 6179234 ----------------CUAACUAUAAUUGGCA-UUGGAAGCCAGAAAUC-----GUUUUUUGCUCUUUCCUUAAAAGCGCUCAAGGUCAC-ACUGC--ACAGAAAAGUGCGGUGUGACCA ----------------..........(((((.-......))))).....-----.......((.((((.....)))).))....((((((-(((((--((......))))))))))))). ( -34.40, z-score = -4.35, R) >droSim1.chrX_random 1822129 95 - 5698898 ----------------CUAACUAUAAUUGGCA-UUGGAAGCCUGAAAUC-----GUUUUUUGCUCUUUCUUUAAAUGCGCUCAAGGUCAC-ACUGC--ACACAAAAGUGUGGUGUGACCA ----------------.............(((-(((((((..(((((..-----...)))))..)))))....)))))......((((((-((..(--((......)))..)))))))). ( -30.80, z-score = -3.65, R) >dp4.chrXL_group1e 10873530 106 - 12523060 ------------CUUACUGCUGUGGGAUCCUUUUCGAAUGCCUGGAUUUUGCCAUUUGGCUGCCCUAUGCUGACUCGCGCGCUGGAUUCCGACUGG--AUGCAUGCGUAUGGUGCUAAGG ------------.........(..((((((.............))))))..)..((((((.(((....((......))((((((.((((.....))--)).)).))))..))))))))). ( -27.12, z-score = 0.83, R) >consensus ________________CUAACUAGAAUUCGCA_UUGAAAGCCUGAAAUC_____GUUUGUUGCACUUUCCUUAAAUGCGCUCAAGGUCAC_ACUGC__ACACAAAAGUGCGGUGUGACCA ....................................................................................((((((.(((((............))))))))))). ( -9.65 = -10.32 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:17:34 2011