
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,279,861 – 5,280,011 |
| Length | 150 |
| Max. P | 0.998921 |

| Location | 5,279,861 – 5,279,960 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 68.26 |
| Shannon entropy | 0.60029 |
| G+C content | 0.38745 |
| Mean single sequence MFE | -18.87 |
| Consensus MFE | -11.24 |
| Energy contribution | -12.63 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.55 |
| SVM RNA-class probability | 0.998921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5279861 99 + 22422827 -------------CCACUAC--UAUAUGUGCCAUAUAAAUAUUCCCCACUUUCAUUUCAUUUUCGACUCAGAUGAGUGACACGUAUUGAUUUGCGUGUCACAUCCUCCGCUCGA -------------.......--((((((...)))))).........................((((..(.((.((((((((((((......)))))))))).)).)).).)))) ( -21.90, z-score = -3.12, R) >droEre2.scaffold_4690 2637200 99 + 18748788 -------------CCACUAC--UAUGUACGCCAUAUAAAUAUUCCCCACUUUCAUUUCAUUUUUGCCUCAGAUGAGUGACACGUAUUGAUUUGCGUGUCACAUCCUCCGCUCGA -------------.......--.......((.........................((((((.......))))))((((((((((......)))))))))).......)).... ( -20.30, z-score = -2.95, R) >droYak2.chrX 18584015 114 - 21770863 CUACAAUGUCACUUCACUGCCAUAUGUAUGCCAUAUAAAUAUUCCCCACUUUCAUUUCAUUUUCGCCUCAGAUGAGUGACACGUAUUGAUUUGCGUGUCACAUCCUCCGCUCGA .(((((((.((......)).))).)))).((.........................((((((.......))))))((((((((((......)))))))))).......)).... ( -22.60, z-score = -2.22, R) >droSec1.super_4 4707386 99 - 6179234 -------------CCACUAC--UAUAUGUGCCAUAUAAAUAUUCCCCACUUUCAUUUCAUUUUCGACUCAGAUGAGUGACACGUAUUGAUUUGCGUGUCACAUCCUCCGCUCGA -------------.......--((((((...)))))).........................((((..(.((.((((((((((((......)))))))))).)).)).).)))) ( -21.90, z-score = -3.12, R) >droSim1.chrX_random 1820493 99 + 5698898 -------------CCACUAC--UAUAUGUGCCAUAUAAAUAUUCCCCACUUUCAUUUCAUUUUCGACUCAGAUGAGUGACACGUAUUGAUUUGCGUGUCACAUCCUCCGCUCGA -------------.......--((((((...)))))).........................((((..(.((.((((((((((((......)))))))))).)).)).).)))) ( -21.90, z-score = -3.12, R) >droAna3.scaffold_13117 2733354 75 + 5790199 -----------------------------------UCGAUAUUUAUAAAUAUUAUCUA--UUUUGCUAGGGAUGAGUGACAUUUAUCGAUU-GCAUGUCACAUC-UCCGCUCGA -----------------------------------((((................(((--......)))(((.(((((((((.((.....)-).))))))).))-)))..)))) ( -15.30, z-score = -0.97, R) >dp4.chrXL_group1e 10872805 90 + 12523060 -------------CUCCUACCGCAUAUAUGAUAGAUUCUUAAACAUGAUUCAAAUUCC--UUCGAUUUCUUUCGAUUACCAUGUUUUUUUUU---UAUAAUUUUUUCC------ -------------.................(((((.....(((((((...........--.((((......))))....)))))))....))---)))..........------ ( -8.19, z-score = -1.08, R) >consensus _____________CCACUAC__UAUAUAUGCCAUAUAAAUAUUCCCCACUUUCAUUUCAUUUUCGACUCAGAUGAGUGACACGUAUUGAUUUGCGUGUCACAUCCUCCGCUCGA ......................................................................((.((((((((((((......)))))))))).)).))....... (-11.24 = -12.63 + 1.39)

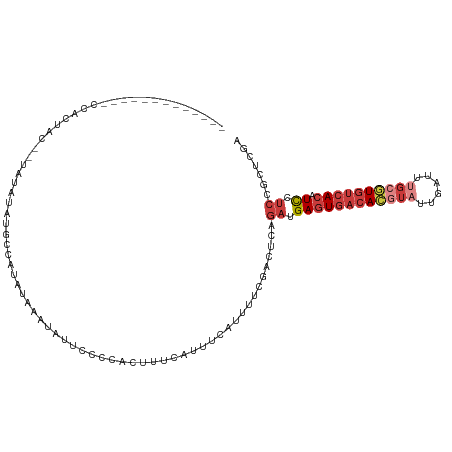
| Location | 5,279,861 – 5,279,960 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 68.26 |
| Shannon entropy | 0.60029 |
| G+C content | 0.38745 |
| Mean single sequence MFE | -22.54 |
| Consensus MFE | -10.70 |
| Energy contribution | -11.99 |
| Covariance contribution | 1.29 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.980031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
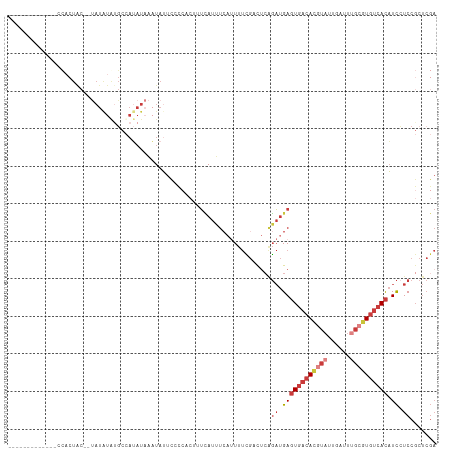
>dm3.chrX 5279861 99 - 22422827 UCGAGCGGAGGAUGUGACACGCAAAUCAAUACGUGUCACUCAUCUGAGUCGAAAAUGAAAUGAAAGUGGGGAAUAUUUAUAUGGCACAUAUA--GUAGUGG------------- ((((.((((.((.((((((((..........)))))))))).))))..))))...((..(((.(((((.....))))).)))..))(((...--...))).------------- ( -24.30, z-score = -1.66, R) >droEre2.scaffold_4690 2637200 99 - 18748788 UCGAGCGGAGGAUGUGACACGCAAAUCAAUACGUGUCACUCAUCUGAGGCAAAAAUGAAAUGAAAGUGGGGAAUAUUUAUAUGGCGUACAUA--GUAGUGG------------- ..(..((((.((.((((((((..........)))))))))).))))...)....(((..(((.(((((.....))))).)))..))).(((.--...))).------------- ( -21.00, z-score = -0.67, R) >droYak2.chrX 18584015 114 + 21770863 UCGAGCGGAGGAUGUGACACGCAAAUCAAUACGUGUCACUCAUCUGAGGCGAAAAUGAAAUGAAAGUGGGGAAUAUUUAUAUGGCAUACAUAUGGCAGUGAAGUGACAUUGUAG (((..((((.((.((((((((..........)))))))))).))))...))).........................((((((.....))))))((((((......)))))).. ( -29.60, z-score = -1.98, R) >droSec1.super_4 4707386 99 + 6179234 UCGAGCGGAGGAUGUGACACGCAAAUCAAUACGUGUCACUCAUCUGAGUCGAAAAUGAAAUGAAAGUGGGGAAUAUUUAUAUGGCACAUAUA--GUAGUGG------------- ((((.((((.((.((((((((..........)))))))))).))))..))))...((..(((.(((((.....))))).)))..))(((...--...))).------------- ( -24.30, z-score = -1.66, R) >droSim1.chrX_random 1820493 99 - 5698898 UCGAGCGGAGGAUGUGACACGCAAAUCAAUACGUGUCACUCAUCUGAGUCGAAAAUGAAAUGAAAGUGGGGAAUAUUUAUAUGGCACAUAUA--GUAGUGG------------- ((((.((((.((.((((((((..........)))))))))).))))..))))...((..(((.(((((.....))))).)))..))(((...--...))).------------- ( -24.30, z-score = -1.66, R) >droAna3.scaffold_13117 2733354 75 - 5790199 UCGAGCGGA-GAUGUGACAUGC-AAUCGAUAAAUGUCACUCAUCCCUAGCAAAA--UAGAUAAUAUUUAUAAAUAUCGA----------------------------------- (((((((((-...(((((((..-.........)))))))...)))...)).(((--((.....))))).......))))----------------------------------- ( -16.70, z-score = -2.17, R) >dp4.chrXL_group1e 10872805 90 - 12523060 ------GGAAAAAAUUAUA---AAAAAAAAACAUGGUAAUCGAAAGAAAUCGAA--GGAAUUUGAAUCAUGUUUAAGAAUCUAUCAUAUAUGCGGUAGGAG------------- ------.............---......(((((((((..((....))..(((((--....)))))))))))))).....((((((........))))))..------------- ( -17.60, z-score = -3.76, R) >consensus UCGAGCGGAGGAUGUGACACGCAAAUCAAUACGUGUCACUCAUCUGAGGCGAAAAUGAAAUGAAAGUGGGGAAUAUUUAUAUGGCACAUAUA__GUAGUGG_____________ .....((((.((.((((((((..........)))))))))).)))).................................................................... (-10.70 = -11.99 + 1.29)


| Location | 5,279,881 – 5,279,998 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 85.75 |
| Shannon entropy | 0.27425 |
| G+C content | 0.37913 |
| Mean single sequence MFE | -21.43 |
| Consensus MFE | -17.81 |
| Energy contribution | -17.51 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.985651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
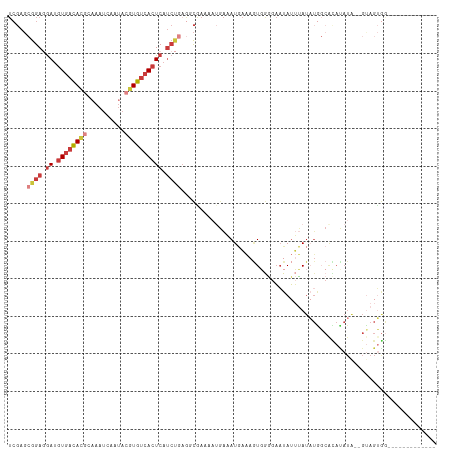
>dm3.chrX 5279881 117 + 22422827 UAAAUAUUCCCCACUUUCAUUUCAUUUUCGACUCAGAUGAGUGACACGUAUUGAUUUGCGUGUCACAUCCUCCGCUCGAUUUGUUUCUGAUUUUCCUUUCCCACGAUUUCAUUACUU ................(((...((...((((..(.((.((((((((((((......)))))))))).)).)).).))))..))....)))........................... ( -22.50, z-score = -3.13, R) >droEre2.scaffold_4690 2637220 117 + 18748788 UAAAUAUUCCCCACUUUCAUUUCAUUUUUGCCUCAGAUGAGUGACACGUAUUGAUUUGCGUGUCACAUCCUCCGCUCGAUUUGUUUCUGAUUUUGCUUUCCCACGAUUUCAUUACUU ...................................(((((((((((((((......)))))))))).......((..((((.......))))..))............))))).... ( -22.30, z-score = -3.04, R) >droYak2.chrX 18584050 117 - 21770863 UAAAUAUUCCCCACUUUCAUUUCAUUUUCGCCUCAGAUGAGUGACACGUAUUGAUUUGCGUGUCACAUCCUCCGCUCGAUUUGUUUCUGAUUUUGCUUUCCCACGAUUUCAUUACUU ...................................(((((((((((((((......)))))))))).......((..((((.......))))..))............))))).... ( -22.30, z-score = -2.93, R) >droSec1.super_4 4707406 117 - 6179234 UAAAUAUUCCCCACUUUCAUUUCAUUUUCGACUCAGAUGAGUGACACGUAUUGAUUUGCGUGUCACAUCCUCCGCUCGAUUUGUUUCUGAUUUUGCUUUCCCACGAUUUCAUUACUU ......................((...((((..(.((.((((((((((((......)))))))))).)).)).).))))..))....(((..(((........)))..)))...... ( -23.40, z-score = -3.00, R) >droSim1.chrX_random 1820513 117 + 5698898 UAAAUAUUCCCCACUUUCAUUUCAUUUUCGACUCAGAUGAGUGACACGUAUUGAUUUGCGUGUCACAUCCUCCGCUCGAUUUGUUUCUGAUUUUGCUUUCCCACGAUUUCAUUACUU ......................((...((((..(.((.((((((((((((......)))))))))).)).)).).))))..))....(((..(((........)))..)))...... ( -23.40, z-score = -3.00, R) >droAna3.scaffold_13117 2733364 102 + 5790199 ----------UAAAUAUUAUCUA--UUUUGCUAGGGAUGAGUGACAUUUAUCGAUU-GCAUGUCACAUC-UCCGCUCGAUUUAUUUUUGAUUUCCGUUUCCGUUUCCAUUCUGAUU- ----------.............--........((((((.(((((((.((.....)-).)))))))...-.....((((.......))))................))))))....- ( -14.70, z-score = 0.50, R) >consensus UAAAUAUUCCCCACUUUCAUUUCAUUUUCGACUCAGAUGAGUGACACGUAUUGAUUUGCGUGUCACAUCCUCCGCUCGAUUUGUUUCUGAUUUUGCUUUCCCACGAUUUCAUUACUU ................(((........(((.....((.((((((((((((......)))))))))).)).))....)))........)))........................... (-17.81 = -17.51 + -0.30)


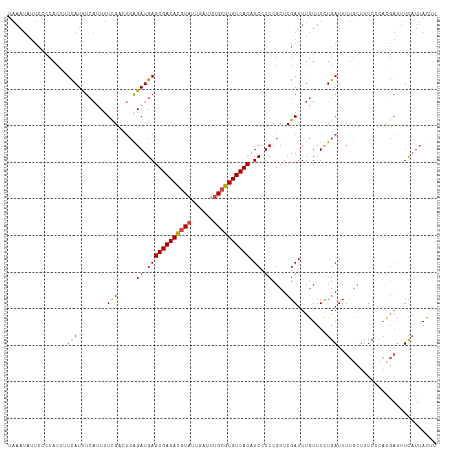
| Location | 5,279,920 – 5,280,011 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 99.56 |
| Shannon entropy | 0.00793 |
| G+C content | 0.39560 |
| Mean single sequence MFE | -21.30 |
| Consensus MFE | -21.20 |
| Energy contribution | -21.40 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.42 |
| Structure conservation index | 1.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.997560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5279920 91 + 22422827 AGUGACACGUAUUGAUUUGCGUGUCACAUCCUCCGCUCGAUUUGUUUCUGAUUUUCCUUUCCCACGAUUUCAUUACUUGGCAUUAUUUUUG .((((((((((......)))))))))).......((.(((...((...(((...((.........))..)))..))))))).......... ( -18.90, z-score = -2.91, R) >droEre2.scaffold_4690 2637259 91 + 18748788 AGUGACACGUAUUGAUUUGCGUGUCACAUCCUCCGCUCGAUUUGUUUCUGAUUUUGCUUUCCCACGAUUUCAUUACUUGGCAUUAUUUUUG .((((((((((......)))))))))).......((.(((...((...(((..(((........)))..)))..))))))).......... ( -21.90, z-score = -3.54, R) >droYak2.chrX 18584089 91 - 21770863 AGUGACACGUAUUGAUUUGCGUGUCACAUCCUCCGCUCGAUUUGUUUCUGAUUUUGCUUUCCCACGAUUUCAUUACUUGGCAUUAUUUUUG .((((((((((......)))))))))).......((.(((...((...(((..(((........)))..)))..))))))).......... ( -21.90, z-score = -3.54, R) >droSec1.super_4 4707445 91 - 6179234 AGUGACACGUAUUGAUUUGCGUGUCACAUCCUCCGCUCGAUUUGUUUCUGAUUUUGCUUUCCCACGAUUUCAUUACUUGGCAUUAUUUUUG .((((((((((......)))))))))).......((.(((...((...(((..(((........)))..)))..))))))).......... ( -21.90, z-score = -3.54, R) >droSim1.chrX_random 1820552 91 + 5698898 AGUGACACGUAUUGAUUUGCGUGUCACAUCCUCCGCUCGAUUUGUUUCUGAUUUUGCUUUCCCACGAUUUCAUUACUUGGCAUUAUUUUUG .((((((((((......)))))))))).......((.(((...((...(((..(((........)))..)))..))))))).......... ( -21.90, z-score = -3.54, R) >consensus AGUGACACGUAUUGAUUUGCGUGUCACAUCCUCCGCUCGAUUUGUUUCUGAUUUUGCUUUCCCACGAUUUCAUUACUUGGCAUUAUUUUUG .((((((((((......)))))))))).......((.(((...((...(((..(((........)))..)))..))))))).......... (-21.20 = -21.40 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:17:31 2011