
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,279,230 – 5,279,323 |
| Length | 93 |
| Max. P | 0.989870 |

| Location | 5,279,230 – 5,279,323 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 81.45 |
| Shannon entropy | 0.33679 |
| G+C content | 0.29978 |
| Mean single sequence MFE | -19.95 |
| Consensus MFE | -12.98 |
| Energy contribution | -13.02 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.596195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
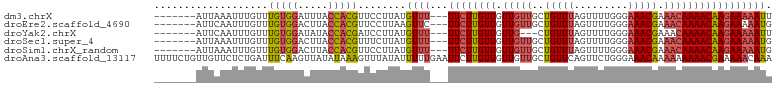
>dm3.chrX 5279230 93 + 22422827 -------AUUAAAUUUGUUUGUGGAUUUACCACGUUCCUUAUGUUU---UUCUUGUUGUUGUUGCUGUUUAGUUUUGGGAAACGAAACAAAACAAGAAAAAUU -------.........(..(((((.....)))))..).....((((---((((((((.(((((..(((((.........))))).))))))))))))))))). ( -21.40, z-score = -2.16, R) >droEre2.scaffold_4690 2636580 93 + 18748788 -------AUUCAAUUUGUUUGUGGACUUACCACGUUCCUUAAGUUC---UUCUUGUUGUUGUUGCUGUUUAGUUUUGGGAAACGAAACAAAACAAGAAAAAUG -------.........(..(((((.....)))))..).....(((.---((((((((.(((((..(((((.........))))).))))))))))))).))). ( -18.60, z-score = -0.70, R) >droYak2.chrX 18583365 90 - 21770863 -------AUUCAAUUUGUUUGUGGAUAUACCACGAUCCUUAUGUUU---UUCUUGUUGUUG---CUGUUUAGUUUUGGGAAACGAAACAAAACAAGAAAAAUU -------...........((((((.....)))))).......((((---((((((((....---.(((((.((((....)))).))))).)))))))))))). ( -21.10, z-score = -2.25, R) >droSec1.super_4 4706769 93 - 6179234 -------AUUAAAUUUGUUUGUGGACUUACCACGUUUCUUAUGUUU---UUCUUGUUGUUGUUGCUGUUUAGUUUUGGGAAACGAAACAAAACAAGAAAAAUG -------.............((((.....)))).........((((---((((((((.(((((..(((((.........))))).))))))))))))))))). ( -21.50, z-score = -2.13, R) >droSim1.chrX_random 1819881 93 + 5698898 -------AUUAAAUUUGUUUGUGGACUUACCACGUUCCUUAUGUUU---UUCUUGUUGUUGUUGCUGUUUAGUUUUGGGAAACGAAACAAAACAAGAAAAAUG -------.........(..(((((.....)))))..).....((((---((((((((.(((((..(((((.........))))).))))))))))))))))). ( -22.00, z-score = -2.23, R) >droAna3.scaffold_13117 2732544 103 + 5790199 UUUUCUGUUGUUCUCUGAUUUCAAGUUAUAUAAAGUUUAUAUUUUUGAAUUCUUGUUGUUGUUGCUGUUCAGUUCUGGGAAACAAAAAAAAACGAAAAACAAA .....((((.(((...((.((((((..(((((.....))))).)))))).))..(((.(((((....((((....)))).))))).....)))))).)))).. ( -15.10, z-score = -0.17, R) >consensus _______AUUAAAUUUGUUUGUGGACUUACCACGUUCCUUAUGUUU___UUCUUGUUGUUGUUGCUGUUUAGUUUUGGGAAACGAAACAAAACAAGAAAAAUG ...................(((((.....)))))...............((((((((((....))......((((((.....))))))..))))))))..... (-12.98 = -13.02 + 0.03)
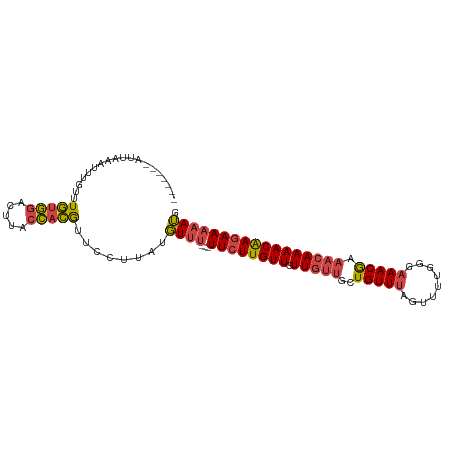
| Location | 5,279,230 – 5,279,323 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 81.45 |
| Shannon entropy | 0.33679 |
| G+C content | 0.29978 |
| Mean single sequence MFE | -18.37 |
| Consensus MFE | -13.12 |
| Energy contribution | -12.51 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.989870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
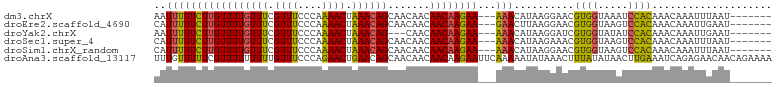
>dm3.chrX 5279230 93 - 22422827 AAUUUUUCUUGUUUUGUUUCGUUUCCCAAAACUAAACAGCAACAACAACAAGAA---AAACAUAAGGAACGUGGUAAAUCCACAAACAAAUUUAAU------- ..(((((((((((((((((.((((....)))).)))))).......))))))))---)))..........((((.....)))).............------- ( -18.61, z-score = -3.21, R) >droEre2.scaffold_4690 2636580 93 - 18748788 CAUUUUUCUUGUUUUGUUUCGUUUCCCAAAACUAAACAGCAACAACAACAAGAA---GAACUUAAGGAACGUGGUAAGUCCACAAACAAAUUGAAU------- ..(((((((((((((((((.((((....)))).)))))).......))))))))---)))..........((((.....)))).............------- ( -18.71, z-score = -1.78, R) >droYak2.chrX 18583365 90 + 21770863 AAUUUUUCUUGUUUUGUUUCGUUUCCCAAAACUAAACAG---CAACAACAAGAA---AAACAUAAGGAUCGUGGUAUAUCCACAAACAAAUUGAAU------- ..(((((((((((((((((.((((....)))).))))))---....))))))))---)))..........((((.....)))).............------- ( -19.60, z-score = -3.31, R) >droSec1.super_4 4706769 93 + 6179234 CAUUUUUCUUGUUUUGUUUCGUUUCCCAAAACUAAACAGCAACAACAACAAGAA---AAACAUAAGAAACGUGGUAAGUCCACAAACAAAUUUAAU------- ..(((((((((((((((((.((((....)))).)))))).......))))))))---)))..........((((.....)))).............------- ( -18.61, z-score = -3.27, R) >droSim1.chrX_random 1819881 93 - 5698898 CAUUUUUCUUGUUUUGUUUCGUUUCCCAAAACUAAACAGCAACAACAACAAGAA---AAACAUAAGGAACGUGGUAAGUCCACAAACAAAUUUAAU------- ..(((((((((((((((((.((((....)))).)))))).......))))))))---)))..........((((.....)))).............------- ( -18.61, z-score = -2.73, R) >droAna3.scaffold_13117 2732544 103 - 5790199 UUUGUUUUUCGUUUUUUUUUGUUUCCCAGAACUGAACAGCAACAACAACAAGAAUUCAAAAAUAUAAACUUUAUAUAACUUGAAAUCAGAGAACAACAGAAAA ((((((.(((((((..(((((.....)))))..))))..............((.(((((..(((((.....)))))...))))).))...))).))))))... ( -16.10, z-score = -1.61, R) >consensus CAUUUUUCUUGUUUUGUUUCGUUUCCCAAAACUAAACAGCAACAACAACAAGAA___AAACAUAAGGAACGUGGUAAAUCCACAAACAAAUUUAAU_______ .....((((((((((((((.((((....)))).)))))).......))))))))................((((.....)))).................... (-13.12 = -12.51 + -0.61)
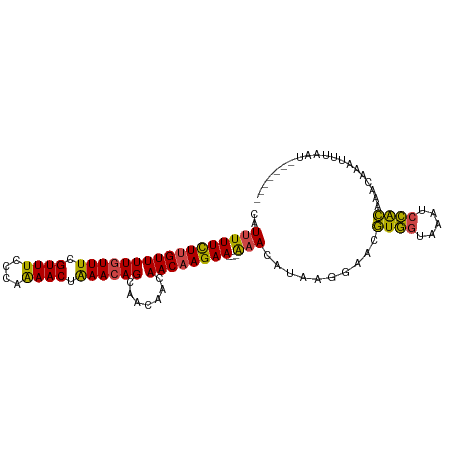
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:17:28 2011