| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,269,360 – 5,269,452 |
| Length | 92 |
| Max. P | 0.980316 |

| Location | 5,269,360 – 5,269,452 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 72.86 |
| Shannon entropy | 0.44932 |
| G+C content | 0.37526 |
| Mean single sequence MFE | -18.82 |
| Consensus MFE | -14.42 |
| Energy contribution | -14.54 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.980316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
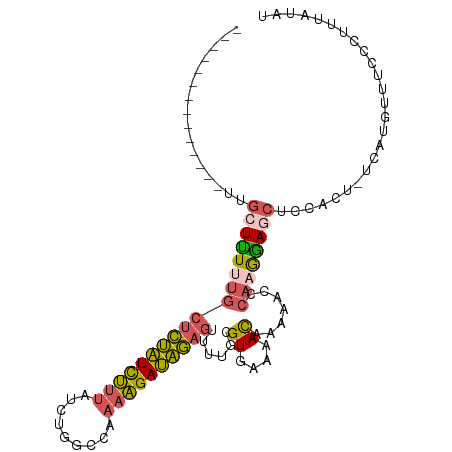
>dm3.chrX 5269360 92 - 22422827 --------------UUGGUUUUUGCUCUAUCUUUAUCUGGCCAAAAGAUAGAGUUUUGCGUGAAAACAAAAACCCAAGGAGCUCCACU-UCAUGUUUCCCUCUAUAU --------------..((((((((((((((((((.........))))))))))).....((....)))))))))...((((....((.-....))....)))).... ( -21.70, z-score = -2.20, R) >droPer1.super_14 305173 103 - 2168203 AUCCGCUCUCUCACUCUAUCUCCAUCUCUUUCUCAAUUGAAAAAAGGAAGAAUUUUUGCGUGAAAAUAAAACU----AGAGUCCCACUACCACAGCCCCCUUUAUCU ............((((((........(((((((...((....)))))))))((((((....)))))).....)----)))))......................... ( -11.00, z-score = -0.12, R) >droEre2.scaffold_4690 2626801 92 - 18748788 --------------UUGCUUUUUGCUCUAUCUGUAUCUGGCUAAAAGAUAGAGUUUUGCGUGAAAACAAAAACCCAAGGAGCUCCACU-UCAUGUUUCCCUUUCUAU --------------..((((((((.(((((((.............)))))))(((((..((....)).))))).))))))))......-.................. ( -18.62, z-score = -1.25, R) >droYak2.chrX 18573561 92 + 21770863 --------------UUGCUUUCUGCUCUAUCUUUAUCUGGGUAAAAGAUAGAGUUUUGCGUGAAAACAAAAACCCAAGAAGCUCCACU-UCAUGGUUCCCUUUAUAU --------------..((.....(((((((((((((....)).)))))))))))...))((....))......(((.((((.....))-)).)))............ ( -22.90, z-score = -2.75, R) >droSec1.super_4 4697013 92 + 6179234 --------------UUGCUUUUUGCUCUAUCUUUAUCUGGCCAAAAGAUAGAGUUUUACGUGAAAACAAAAACCCAAGGAGCUCCACU-UCAUGUUUUCCUUUAUAU --------------..((((((((((((((((((.........))))))))))......((....)).......))))))))......-.................. ( -19.90, z-score = -2.41, R) >consensus ______________UUGCUUUUUGCUCUAUCUUUAUCUGGCCAAAAGAUAGAGUUUUGCGUGAAAACAAAAACCCAAGGAGCUCCACU_UCAUGUUUCCCUUUAUAU ................((((((((((((((((((.........))))))))))......((....)).......))))))))......................... (-14.42 = -14.54 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:17:26 2011