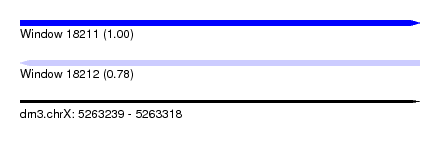
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,263,239 – 5,263,318 |
| Length | 79 |
| Max. P | 0.998018 |

| Location | 5,263,239 – 5,263,318 |
|---|---|
| Length | 79 |
| Sequences | 6 |
| Columns | 83 |
| Reading direction | forward |
| Mean pairwise identity | 74.47 |
| Shannon entropy | 0.48268 |
| G+C content | 0.41488 |
| Mean single sequence MFE | -20.03 |
| Consensus MFE | -13.69 |
| Energy contribution | -14.63 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.24 |
| SVM RNA-class probability | 0.998018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
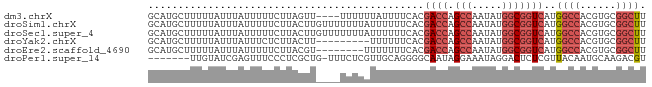
>dm3.chrX 5263239 79 + 22422827 AAGCCGCACGUGGCCAUGACCGCCAUAUUGGCUGGUCGUGAAAAUAAAAAA----AACUAAGAAAAAUAAAUAAAAAGCAUGC ..((((....))))((((((((((.....))).)))))))...........----............................ ( -20.70, z-score = -3.14, R) >droSim1.chrX 4055370 83 + 17042790 AAGCCGCACGUGGCCAUGACCGCCAUAUUGGCUGGUCGUGAAAAAAUAAAAAAACAAGUAAGAAAAAUAAAUAAAAAGCAUGC ..((((....))))((((((((((.....))).)))))))........................................... ( -20.70, z-score = -2.92, R) >droSec1.super_4 4691103 83 - 6179234 AAGCCGCACGUGGCCAUGACCGCCAUAUUGGCUGGUCGUGAAAAAAUAAAAAAACAAGUAAGAAAAAUAAAUAAAAAGCAUGC ..((((....))))((((((((((.....))).)))))))........................................... ( -20.70, z-score = -2.92, R) >droYak2.chrX 18567505 74 - 21770863 AAGCCGCACGUGGCCAUGACCGCCAUAUUGGCUGGUCGUGAAAAAA---------AAGUAAGAGAAAUAAAUAAAAAGCAUGC ..((((....))))((((((((((.....))).)))))))......---------............................ ( -20.70, z-score = -2.86, R) >droEre2.scaffold_4690 2620779 75 + 18748788 AAGCCGCACGUGGCCAUGACCGCCAUAUUGGCUGGUCGUGAAAAAAA--------ACGUAAGAAAAAUAAAUAAAAAGCAUGC ..((((....))))((((((((((.....))).))))))).......--------.(....)..................... ( -20.90, z-score = -2.67, R) >droPer1.super_14 297383 75 + 2168203 ACGUCUUGCAUUGUAACGAGAGUCCUAUUUCCUAUUGCCCCUGCAACGAGAAA-CAGCGAGGGAAACUCGAUACAA------- ..........(((((.((((..((((........((((....))))((.....-...)).))))..)))).)))))------- ( -16.50, z-score = -1.00, R) >consensus AAGCCGCACGUGGCCAUGACCGCCAUAUUGGCUGGUCGUGAAAAAAAAAAA____AAGUAAGAAAAAUAAAUAAAAAGCAUGC ..((((....))))((((((((((.....))).)))))))........................................... (-13.69 = -14.63 + 0.95)

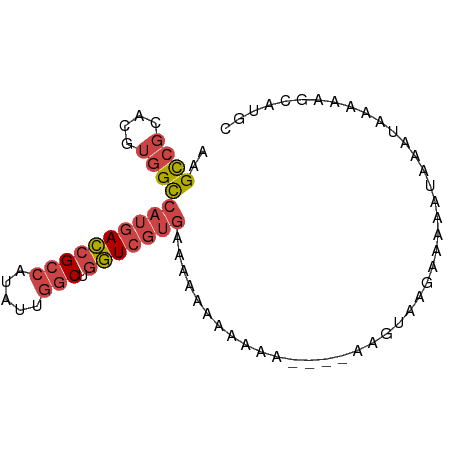
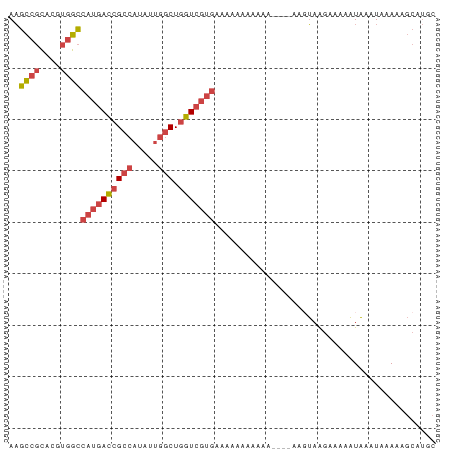
| Location | 5,263,239 – 5,263,318 |
|---|---|
| Length | 79 |
| Sequences | 6 |
| Columns | 83 |
| Reading direction | reverse |
| Mean pairwise identity | 74.47 |
| Shannon entropy | 0.48268 |
| G+C content | 0.41488 |
| Mean single sequence MFE | -17.48 |
| Consensus MFE | -9.86 |
| Energy contribution | -10.92 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.782150 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5263239 79 - 22422827 GCAUGCUUUUUAUUUAUUUUUCUUAGUU----UUUUUUAUUUUCACGACCAGCCAAUAUGGCGGUCAUGGCCACGUGCGGCUU (((((..................(((..----....)))...(((.((((.(((.....))))))).)))...)))))..... ( -16.30, z-score = -1.23, R) >droSim1.chrX 4055370 83 - 17042790 GCAUGCUUUUUAUUUAUUUUUCUUACUUGUUUUUUUAUUUUUUCACGACCAGCCAAUAUGGCGGUCAUGGCCACGUGCGGCUU ..............................................((((.(((.....)))))))..((((......)))). ( -16.10, z-score = -1.15, R) >droSec1.super_4 4691103 83 + 6179234 GCAUGCUUUUUAUUUAUUUUUCUUACUUGUUUUUUUAUUUUUUCACGACCAGCCAAUAUGGCGGUCAUGGCCACGUGCGGCUU ..............................................((((.(((.....)))))))..((((......)))). ( -16.10, z-score = -1.15, R) >droYak2.chrX 18567505 74 + 21770863 GCAUGCUUUUUAUUUAUUUCUCUUACUU---------UUUUUUCACGACCAGCCAAUAUGGCGGUCAUGGCCACGUGCGGCUU ............................---------.........((((.(((.....)))))))..((((......)))). ( -16.10, z-score = -1.47, R) >droEre2.scaffold_4690 2620779 75 - 18748788 GCAUGCUUUUUAUUUAUUUUUCUUACGU--------UUUUUUUCACGACCAGCCAAUAUGGCGGUCAUGGCCACGUGCGGCUU (((((.....................((--------........))((((.(((.....))))))).......)))))..... ( -16.40, z-score = -1.16, R) >droPer1.super_14 297383 75 - 2168203 -------UUGUAUCGAGUUUCCCUCGCUG-UUUCUCGUUGCAGGGGCAAUAGGAAAUAGGACUCUCGUUACAAUGCAAGACGU -------(((((.((((..(((.....((-(((((..((((....))))..))))))))))..)))).))))).......... ( -23.90, z-score = -2.43, R) >consensus GCAUGCUUUUUAUUUAUUUUUCUUACUU____UUUUAUUUUUUCACGACCAGCCAAUAUGGCGGUCAUGGCCACGUGCGGCUU ..............................................((((.(((.....)))))))..((((......)))). ( -9.86 = -10.92 + 1.06)
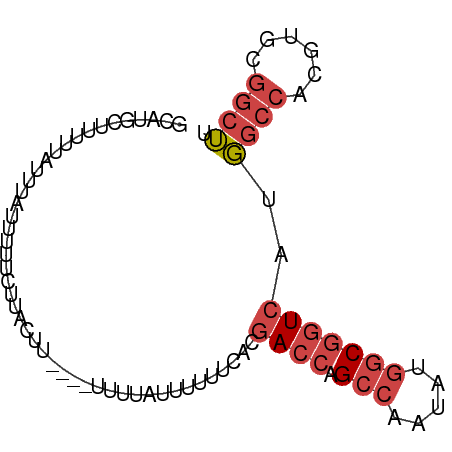
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:17:25 2011