| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,255,360 – 5,255,429 |
| Length | 69 |
| Max. P | 0.771882 |

| Location | 5,255,360 – 5,255,429 |
|---|---|
| Length | 69 |
| Sequences | 7 |
| Columns | 88 |
| Reading direction | reverse |
| Mean pairwise identity | 60.90 |
| Shannon entropy | 0.67913 |
| G+C content | 0.55768 |
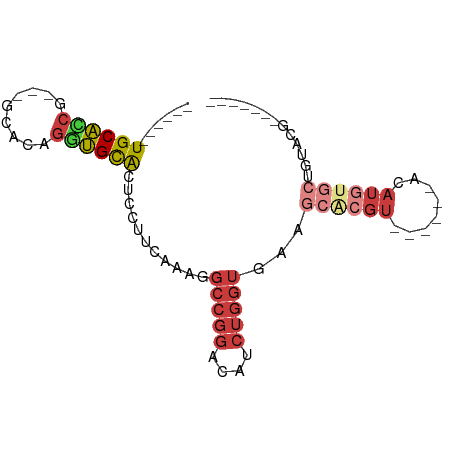
| Mean single sequence MFE | -21.22 |
| Consensus MFE | -11.15 |
| Energy contribution | -12.50 |
| Covariance contribution | 1.35 |
| Combinations/Pair | 1.35 |
| Mean z-score | -0.61 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.771882 |
| Prediction | RNA |
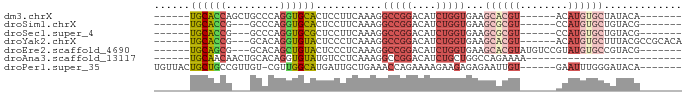
Download alignment: ClustalW | MAF
>dm3.chrX 5255360 69 - 22422827 ------UGCACCAGCUGCCCAGGUGCACUCCUUCAAAGGCCGGACAUCUGGUGAAGCACGU------ACAUGUGCUAUACA------- ------..((((((.((.((.(((..............))))).)).)))))).((((((.------...)))))).....------- ( -21.24, z-score = -0.42, R) >droSim1.chrX 4047927 66 - 17042790 ------UGCACCG---GCCCAGGUGCACUCCUUCAAAGGCCGGACAUCUGGUGAAGCGCGU------CCAUGUGCUGUACG------- ------....(((---(((.(((......))).....)))))).......(((.((((((.------...)))))).))).------- ( -22.80, z-score = -0.48, R) >droSec1.super_4 4683675 66 + 6179234 ------UGCACCG---GCCCAGGUGCGCUCCUUCAAAGGCCGGACAUCUGGUGAAGCGCGU------CCAUGUGCUGUACG------- ------....(((---(((.(((......))).....)))))).......(((.((((((.------...)))))).))).------- ( -22.80, z-score = -0.14, R) >droYak2.chrX 18559880 73 + 21770863 ------UGCACCG---GCACAGGUGUACUCCCUCAAAGGCCGGACAUCUGGUGAAGCACGU------ACAUGUGCUUUACGCCGCACA ------.....((---((.(((((((....((.....))....)))))))((((((((((.------...)))))))))))))).... ( -30.90, z-score = -3.03, R) >droEre2.scaffold_4690 2612868 72 - 18748788 ------UGCAGCG---GCACAGCUGUACUCCCUCAAAGGCCGGACAUCUGGUGAAGCACGUAUGUCCGUAUGUGCCGUACG------- ------....(((---((((((((.............)))(((((((.((........)).)))))))..))))))))...------- ( -25.92, z-score = -1.24, R) >droAna3.scaffold_13117 2704667 56 - 5790199 ------UGCAACAACUGCACAGGUGUAUGUCCUCAAAGGCCGGACAUCUGCUGGCCAGAAAA-------------------------- ------((((.....)))).(((.......)))....((((((.(....)))))))......-------------------------- ( -14.60, z-score = -0.32, R) >droPer1.super_35 759090 74 + 973955 UGUUACUGCUGCCGUUGU-CGUUGGCAUGAUUGCUGAAACCAGAAAAGAAGAGAGAAUUGU------GAAUUUGGGAUACA------- .............((.((-(.(..(((....)))..)..(((((..(.((.......)).)------...)))))))))).------- ( -10.30, z-score = 1.35, R) >consensus ______UGCACCG___GCACAGGUGCACUCCUUCAAAGGCCGGACAUCUGGUGAAGCACGU______ACAUGUGCUGUACG_______ ......((((((.........))))))...........(((((....)))))...((((((........))))))............. (-11.15 = -12.50 + 1.35)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:17:24 2011