
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,252,918 – 5,253,033 |
| Length | 115 |
| Max. P | 0.999885 |

| Location | 5,252,918 – 5,253,033 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 132 |
| Reading direction | forward |
| Mean pairwise identity | 63.89 |
| Shannon entropy | 0.65003 |
| G+C content | 0.32098 |
| Mean single sequence MFE | -24.07 |
| Consensus MFE | -10.03 |
| Energy contribution | -11.42 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.977068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
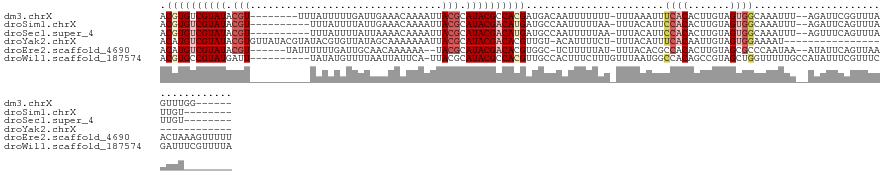
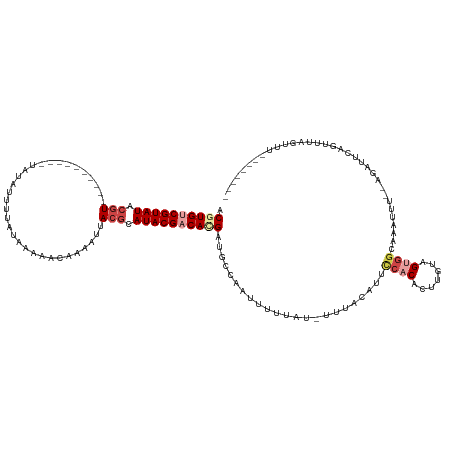
>dm3.chrX 5252918 115 + 22422827 ACGUGUCGUAUACGU--------UUUAUUUUUGAUUGAAACAAAAUUACGCAUACGCCACGAUGACAAUUUUUUU-UUUAAAUUUCACACUUGUAGUGGCAAAUUU--AGAUUCGGUUUAGUUUGG------ .((((.(((((.(((--------.....(((((.......)))))..))).))))).))))..............-(((((((((..(((.....)))..))))))--)))...............------ ( -22.20, z-score = -1.01, R) >droSim1.chrX 4045601 111 + 17042790 ACGUGUCGUAUACGU----------UUUAUUUUAUUGAAACAAAAUUACGCAUACGACAUGAUGCCAAUUUUUAA-UUUACAUUCCACACUUGUAGUGGCAAAUUU--AGAUUCAGUUUAUUGU-------- .((((((((((.(((----------...(((((........))))).))).)))))))))).(((((........-.(((((.........)))))))))).....--................-------- ( -24.00, z-score = -2.41, R) >droSec1.super_4 4681352 111 - 6179234 ACGUGUCGUAUACGU----------UUUAUUUUAUUAAAACAAAAUUACGCAUACGACAUGAUGCCAAUUUUUAA-UUUACAUUCCACACUUGUAGUGGCAAAUUU--AGUUUCAGUUUAUUGU-------- .((((((((((.(((----------...(((((........))))).))).)))))))))).(((((........-.(((((.........)))))))))).....--................-------- ( -24.00, z-score = -2.73, R) >droYak2.chrX 18557520 102 - 21770863 ACAUGUCGUAUACGUGUUAUACGUAUACGUGUUAUAGCAAAAAAAUUACGCAUACGACACGUUGU-ACAUUUUCU-UUUACAUUUCACAAUUGUAGUGGAAAAU---------------------------- .......((((((((((....(((((.((((...............)))).)))))))))).)))-))(((((((-.(((((.........))))).)))))))---------------------------- ( -24.06, z-score = -1.50, R) >droEre2.scaffold_4690 2610407 120 + 18748788 ACAUGUCGUAUACGU------UAUUUUUUGAUUGCAACAAAAAA--UACGCAUACGACACGUGGC-UCUUUUUAU-UUUACACGCCACACUUGUAGCGCCCAAUAA--AUAUUCAGUUAAACUAAAGUUUUU ...((((((((.(((------.((((((((.......)))))))--)))).)))))))).(((((-.........-.......)))))((((.((((.........--.......)))).....)))).... ( -26.78, z-score = -3.07, R) >droWil1.scaffold_187574 101 121 + 1299 ACGUGCCGUAUGAUU----------UAUAUGUUUUAAUUAUUCA-UUACGCAUACGCCACGUUGCCACUUUCUUUGUUUAAUGGCCACAGCCGUAGCUGGUUUUUGCCAUAUUUCGUUUCGAUUUCGUUUUA (((((.((((((...----------...(((..(.....)..))-)....)))))).)))))..................(((((..((((....))))......)))))...................... ( -23.40, z-score = -0.84, R) >consensus ACGUGUCGUAUACGU__________UAUAUUUUAUAAAAACAAAAUUACGCAUACGACACGAUGCCAAUUUUUAU_UUUACAUUCCACACUUGUAGUGGCAAAUUU__AGAUUCAGUUUAGUUU________ .((((((((((.(((................................))).)))))))))).......................((((.......))))................................. (-10.03 = -11.42 + 1.39)
| Location | 5,252,918 – 5,253,033 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 132 |
| Reading direction | reverse |
| Mean pairwise identity | 63.89 |
| Shannon entropy | 0.65003 |
| G+C content | 0.32098 |
| Mean single sequence MFE | -29.12 |
| Consensus MFE | -15.44 |
| Energy contribution | -16.13 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.71 |
| SVM RNA-class probability | 0.999885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
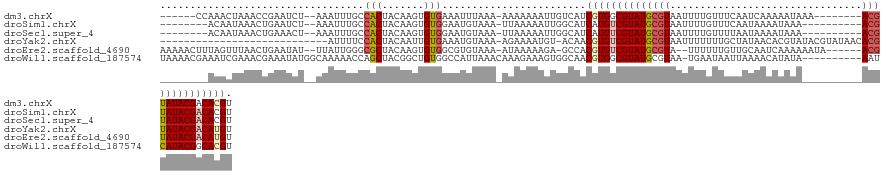
>dm3.chrX 5252918 115 - 22422827 ------CCAAACUAAACCGAAUCU--AAAUUUGCCACUACAAGUGUGAAAUUUAAA-AAAAAAAUUGUCAUCGUGGCGUAUGCGUAAUUUUGUUUCAAUCAAAAAUAAA--------ACGUAUACGACACGU ------.................(--((((((..(((.....)))..)))))))..-..............((((.(((((((((...(((((((.......)))))))--------))))))))).)))). ( -24.20, z-score = -3.02, R) >droSim1.chrX 4045601 111 - 17042790 --------ACAAUAAACUGAAUCU--AAAUUUGCCACUACAAGUGUGGAAUGUAAA-UUAAAAAUUGGCAUCAUGUCGUAUGCGUAAUUUUGUUUCAAUAAAAUAAA----------ACGUAUACGACACGU --------................--.....(((((((((....))))(((....)-))......)))))...((((((((((((.(((((((....)))))))...----------))))))))))))... ( -27.20, z-score = -3.37, R) >droSec1.super_4 4681352 111 + 6179234 --------ACAAUAAACUGAAACU--AAAUUUGCCACUACAAGUGUGGAAUGUAAA-UUAAAAAUUGGCAUCAUGUCGUAUGCGUAAUUUUGUUUUAAUAAAAUAAA----------ACGUAUACGACACGU --------................--.....(((((((((....))))(((....)-))......)))))...((((((((((((.(((((((....)))))))...----------))))))))))))... ( -27.20, z-score = -3.30, R) >droYak2.chrX 18557520 102 + 21770863 ----------------------------AUUUUCCACUACAAUUGUGAAAUGUAAA-AGAAAAUGU-ACAACGUGUCGUAUGCGUAAUUUUUUUGCUAUAACACGUAUACGUAUAACACGUAUACGACAUGU ----------------------------((((((...((((.((....))))))..-.))))))..-...(((((((((((((((((.....))))......(((....))).......))))))))))))) ( -25.50, z-score = -1.90, R) >droEre2.scaffold_4690 2610407 120 - 18748788 AAAAACUUUAGUUUAACUGAAUAU--UUAUUGGGCGCUACAAGUGUGGCGUGUAAA-AUAAAAAGA-GCCACGUGUCGUAUGCGUA--UUUUUUGUUGCAAUCAAAAAAUA------ACGUAUACGACAUGU ......(((((.....)))))...--...(((.(((((((....))))))).))).-.........-...((((((((((((((((--(((((((.......)))))))).------))))))))))))))) ( -41.90, z-score = -5.76, R) >droWil1.scaffold_187574 101 121 - 1299 UAAAACGAAAUCGAAACGAAAUAUGGCAAAAACCAGCUACGGCUGUGGCCAUUAAACAAAGAAAGUGGCAACGUGGCGUAUGCGUAA-UGAAUAAUUAAAACAUAUA----------AAUCAUACGGCACGU .....((....))...................(((((....)))).)((((((..........)))))).(((((.((((((..(((-((...........))).))----------...)))))).))))) ( -28.70, z-score = -2.24, R) >consensus ________AAAAUAAACUGAAUCU__AAAUUUGCCACUACAAGUGUGGAAUGUAAA_AUAAAAAGUGGCAACGUGUCGUAUGCGUAAUUUUGUUUCAAUAAAAUAAA__________ACGUAUACGACACGU ..................................(((.......)))........................((((((((((((((................................)))))))))))))). (-15.44 = -16.13 + 0.70)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:17:23 2011