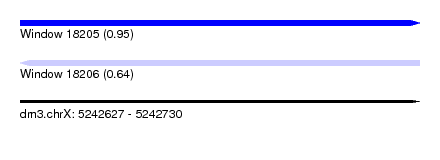
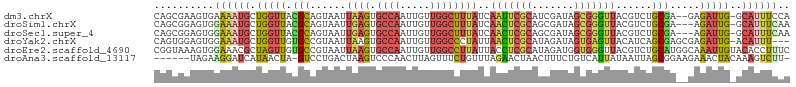
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,242,627 – 5,242,730 |
| Length | 103 |
| Max. P | 0.945237 |

| Location | 5,242,627 – 5,242,730 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 66.05 |
| Shannon entropy | 0.65192 |
| G+C content | 0.43969 |
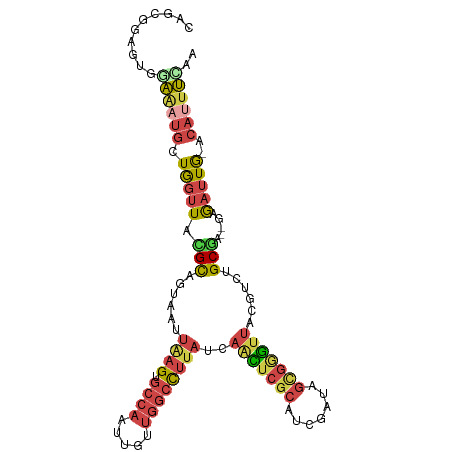
| Mean single sequence MFE | -23.71 |
| Consensus MFE | -10.28 |
| Energy contribution | -9.93 |
| Covariance contribution | -0.35 |
| Combinations/Pair | 1.65 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.945237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
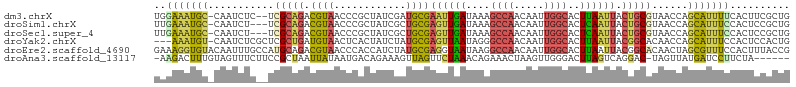
>dm3.chrX 5242627 103 + 22422827 UGGAAAUGC-CAAUCUC--UCGCAGACGUAACCCGCUAUCGAUGCGAAUUGAUAAAGCCAACAAUUGGCACUUAAUUACUGCGUAACCAGCAUUUUCACUUCGCUG (((((((((-.......--.(((((...........(((((((....)))))))..((((.....)))).........)))))......)))))))))........ ( -27.34, z-score = -2.96, R) >droSim1.chrX 4034989 102 + 17042790 UUGAAAUGC-CAAUCU---UCGCAGACGUAACCCGCUAUCGCUGCGAGUUGAUAAAGCCAACAAUUGGCACUCAAUUACUGCGUAACCAGCAUUUCCACUCCGCUG ..(((((((-......---.(((((...((((.(((.......))).)))).....((((.....)))).........)))))......))))))).......... ( -27.62, z-score = -3.04, R) >droSec1.super_4 4670404 102 - 6179234 UUGAAAUGC-CAAUCU---UCGCAGACGUAACCCGCUAUCGCUGCGAGUUGAUAAAGCCAACAAUUGGCACUCAAUUACUGCGUAACCAGCAUUUCCACUCCGCUG ..(((((((-......---.(((((...((((.(((.......))).)))).....((((.....)))).........)))))......))))))).......... ( -27.62, z-score = -3.04, R) >droYak2.chrX 18546420 102 - 21770863 ---AAAUGU-CAAUCUCGCUCGCUGAUGUAACUCACUAUCUAUGCGAGUUAAUAGGGCCAACAAUUGGCACUUAAUUACGGCACAACCAGCAUUUCCACUCCACUG ---...(((-(......((((((.((((........))))...))))))...((((((((.....)))).)))).....))))....................... ( -22.00, z-score = -1.87, R) >droEre2.scaffold_4690 2597079 106 + 18748788 GAAAGGUGUACAAUUUGCCAUGCAGACGUAACCCACCAUCUAUGCGAGGUAAUAAGGCCAACAAUUGGCACUUAAUUACGGCACAACUAGCGUUUCCACUUUACCG ((((.((........((((.((((..................))))..((((((((((((.....)))).))).)))))))))......)).)))).......... ( -22.71, z-score = -0.41, R) >droAna3.scaffold_13117 2687115 98 + 5790199 -AAGACUUUGUAGUUUCUUCCGCUAAUUAUAAUGACAGAAAGUUAGUUCUAAACAGAAACUAAGUUGGGACUUAGUCAGGAC-UAGUUAUGAUCCUUCUA------ -..((((...((((((((...(((((((............))))))).......))))))))((((..(((...)))..)))-)))))............------ ( -15.00, z-score = 1.51, R) >consensus UAGAAAUGC_CAAUCUC__UCGCAGACGUAACCCACUAUCGAUGCGAGUUAAUAAAGCCAACAAUUGGCACUUAAUUACGGCGUAACCAGCAUUUCCACUCCGCUG ..(((((((...........(((((.((((............))))((((((....((((.....))))..)))))).)))))......))))))).......... (-10.28 = -9.93 + -0.35)
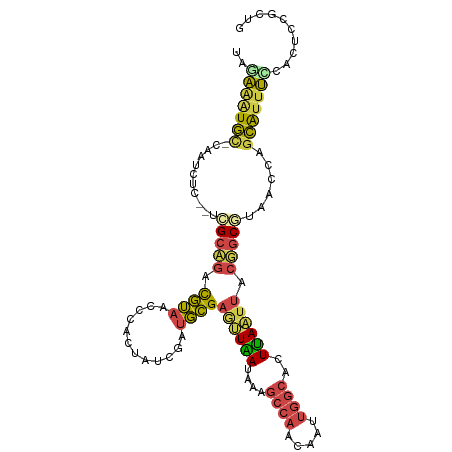
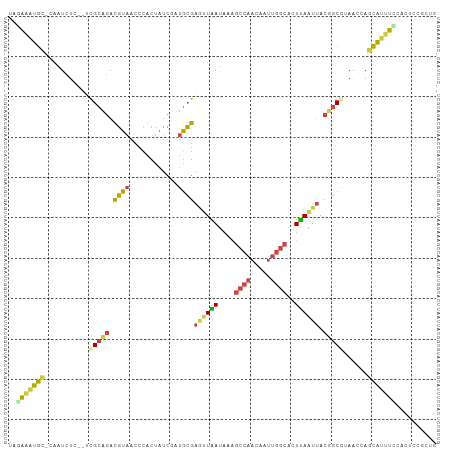
| Location | 5,242,627 – 5,242,730 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 66.05 |
| Shannon entropy | 0.65192 |
| G+C content | 0.43969 |
| Mean single sequence MFE | -29.93 |
| Consensus MFE | -10.58 |
| Energy contribution | -11.08 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.640586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5242627 103 - 22422827 CAGCGAAGUGAAAAUGCUGGUUACGCAGUAAUUAAGUGCCAAUUGUUGGCUUUAUCAAUUCGCAUCGAUAGCGGGUUACGUCUGCGA--GAGAUUG-GCAUUUCCA ........((.((((((..(((.(((((....((((.((((.....))))))))..(((((((.......)))))))....))))).--..)))..-)))))).)) ( -29.10, z-score = -0.91, R) >droSim1.chrX 4034989 102 - 17042790 CAGCGGAGUGGAAAUGCUGGUUACGCAGUAAUUGAGUGCCAAUUGUUGGCUUUAUCAACUCGCAGCGAUAGCGGGUUACGUCUGCGA---AGAUUG-GCAUUUCAA ..........(((((((..(((.(((((.....((..((((.....))))....))(((((((.......)))))))....))))).---.)))..-))))))).. ( -35.90, z-score = -2.40, R) >droSec1.super_4 4670404 102 + 6179234 CAGCGGAGUGGAAAUGCUGGUUACGCAGUAAUUGAGUGCCAAUUGUUGGCUUUAUCAACUCGCAGCGAUAGCGGGUUACGUCUGCGA---AGAUUG-GCAUUUCAA ..........(((((((..(((.(((((.....((..((((.....))))....))(((((((.......)))))))....))))).---.)))..-))))))).. ( -35.90, z-score = -2.40, R) >droYak2.chrX 18546420 102 + 21770863 CAGUGGAGUGGAAAUGCUGGUUGUGCCGUAAUUAAGUGCCAAUUGUUGGCCCUAUUAACUCGCAUAGAUAGUGAGUUACAUCAGCGAGCGAGAUUG-ACAUUU--- .....(((((..((((((.((((((.........((.((((.....)))).))..((((((((.......)))))))))).)))).)))...))).-.)))))--- ( -27.20, z-score = -1.03, R) >droEre2.scaffold_4690 2597079 106 - 18748788 CGGUAAAGUGGAAACGCUAGUUGUGCCGUAAUUAAGUGCCAAUUGUUGGCCUUAUUACCUCGCAUAGAUGGUGGGUUACGUCUGCAUGGCAAAUUGUACACCUUUC .(((..((((....)))).((..(((((((((((((.((((.....))))))))(((((((.....)).)))))))))))...)))..)).........))).... ( -34.50, z-score = -2.20, R) >droAna3.scaffold_13117 2687115 98 - 5790199 ------UAGAAGGAUCAUAACUA-GUCCUGACUAAGUCCCAACUUAGUUUCUGUUUAGAACUAACUUUCUGUCAUUAUAAUUAGCGGAAGAAACUACAAAGUCUU- ------....(((((........-)))))((((((((....))))((((((((((.......)))..((((..(.......)..)))))))))))....))))..- ( -17.00, z-score = 0.01, R) >consensus CAGCGGAGUGGAAAUGCUGGUUACGCAGUAAUUAAGUGCCAAUUGUUGGCCUUAUCAACUCGCAUCGAUAGCGGGUUACGUCUGCGA__GAGAUUG_ACAUUUCAA ..((..((((....)))).....(((......((((.((((.....))))))))..(((((((.......)))))))......)))...........))....... (-10.58 = -11.08 + 0.51)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:17:20 2011