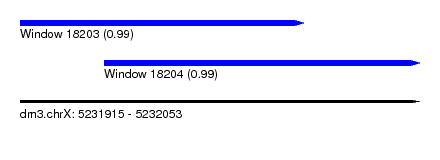
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,231,915 – 5,232,053 |
| Length | 138 |
| Max. P | 0.993444 |

| Location | 5,231,915 – 5,232,013 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 71.24 |
| Shannon entropy | 0.44719 |
| G+C content | 0.35724 |
| Mean single sequence MFE | -25.11 |
| Consensus MFE | -19.19 |
| Energy contribution | -20.00 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.989608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
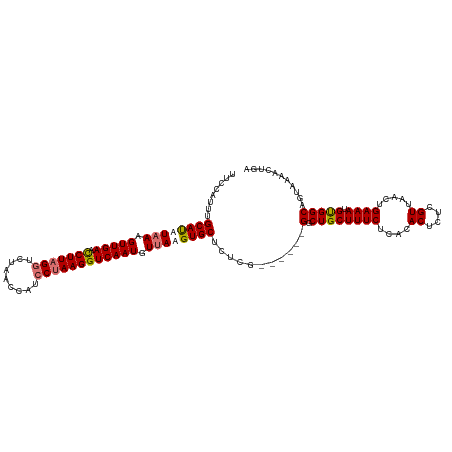
>dm3.chrX 5231915 98 + 22422827 UCCACUAUAUAUUUUAAUAUGUGCAAUAGAUU---------------UGCAUUUAAGGUUGAAUCUUAGGUCUAACUAUCCUAAGGUCAAUGUUAAGUGCUUUCG------GGCUGCUU (((..(((((((....))))))).....((..---------------.((((((((.(((((.(((((((.........)))))))))))).))))))))..)))------))...... ( -28.30, z-score = -3.53, R) >droYak2.chrX 18536673 118 - 21770863 -GCUAUAAACAAUGCAAUAUGUGUAAUAGAUUGUAAGCUUUUCCAUUUGCAUAUAAAGUUGAACCUUAGGUCUAACCAUCCUAAGGUCAAUGUUAAGUGCUUUCUCUGCUCGUCUGCUU -((.........((((((.(((...))).))))))(((..........((((.(((.(((((.(((((((.........)))))))))))).))).)))).......))).....)).. ( -25.13, z-score = -1.63, R) >droEre2.scaffold_4690 2587275 101 + 18748788 --UCCAUAAGAUUGAAGUAUGUGUAAGCUUUU----------CCAUUUGCACAUUAAGUUGAACCUUCGCUCUUACCAUCCUAAGGUCAAUGUUAAGUGCUCCUC------GGCUGCUU --............(((((((((((((.....----------...)))))))))..((((((......((.((((.(((((...))...))).)))).))...))------)))))))) ( -16.80, z-score = 0.38, R) >droSec1.super_4 4661037 112 - 6179234 UUCACAAUAUGUUUAAAUAUGUGUAAUAGAUUGUUAGCUUUUGCAUUUGCAUUUAAGGUUGAAUCUUAGGUCUAACUAUCCUAAGGUCAAUGUUAAGUGCUCUCG------G-CUGCUU ..((((.(((......)))))))............(((....((....((((((((.(((((.(((((((.........)))))))))))).)))))))).....------)-).))). ( -30.20, z-score = -3.21, R) >consensus __CACAAAAUAUUGAAAUAUGUGUAAUAGAUU__________CCAUUUGCAUAUAAAGUUGAACCUUAGGUCUAACCAUCCUAAGGUCAAUGUUAAGUGCUCUCG______GGCUGCUU ................................................((((((((.(((((.(((((((.........)))))))))))).))))))))................... (-19.19 = -20.00 + 0.81)


| Location | 5,231,944 – 5,232,053 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 85.49 |
| Shannon entropy | 0.23037 |
| G+C content | 0.40182 |
| Mean single sequence MFE | -29.44 |
| Consensus MFE | -23.85 |
| Energy contribution | -24.97 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.993444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5231944 109 + 22422827 ----AUUUGCAUUUAAGGUUGAAUCUUAGGUCUAACUAUCCUAAGGUCAAUGUUAAGUGCUUUCG------GGCUGCUUUCUCACACUCUCGUUAACUGAAAUGUGGCAGUAAAACUGA ----....((((((((.(((((.(((((((.........)))))))))))).))))))))..(((------((((((.....((((...(((.....)))..)))))))))....)))) ( -31.70, z-score = -3.48, R) >droYak2.chrX 18536712 119 - 21770863 UUCCAUUUGCAUAUAAAGUUGAACCUUAGGUCUAACCAUCCUAAGGUCAAUGUUAAGUGCUUUCUCUGCUCGUCUGCUUUCUCCUACUAUCGUUAACUGAAAUGUGGCAGUAAAACUGA ........((((.(((.(((((.(((((((.........)))))))))))).))).))))......((((.(((.((((((....((....)).....)))).)))))))))....... ( -26.90, z-score = -2.57, R) >droEre2.scaffold_4690 2587303 113 + 18748788 UUCCAUUUGCACAUUAAGUUGAACCUUCGCUCUUACCAUCCUAAGGUCAAUGUUAAGUGCUCCUC------GGCUGCUUUCUCAUACUCUCGUUAACUGAAAUGCGGCCGUAAAACUGA ........((((.(((((((((.((((...............))))))))).))))))))....(------((((((((((....((....)).....)))).)))))))......... ( -26.66, z-score = -2.66, R) >droSec1.super_4 4661077 112 - 6179234 UUGCAUUUGCAUUUAAGGUUGAAUCUUAGGUCUAACUAUCCUAAGGUCAAUGUUAAGUGCUCUCG------G-CUGCUUUCUCGCACUCUCGUUAACUGAAAUGUGGCAGUAAAACUGA (..(((((((((((((.(((((.(((((((.........)))))))))))).))))))))....(------.-.(((......)))..)..........)))))..)(((.....))). ( -32.50, z-score = -3.24, R) >consensus UUCCAUUUGCAUAUAAAGUUGAACCUUAGGUCUAACCAUCCUAAGGUCAAUGUUAAGUGCUCUCG______GGCUGCUUUCUCACACUCUCGUUAACUGAAAUGUGGCAGUAAAACUGA ........((((((((.(((((.(((((((.........)))))))))))).))))))))............(((((((((....((....)).....)))).)))))........... (-23.85 = -24.97 + 1.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:17:19 2011