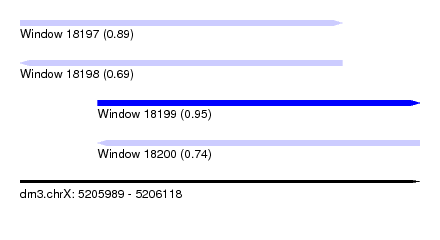
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,205,989 – 5,206,118 |
| Length | 129 |
| Max. P | 0.953853 |

| Location | 5,205,989 – 5,206,093 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 71.05 |
| Shannon entropy | 0.47608 |
| G+C content | 0.46843 |
| Mean single sequence MFE | -33.50 |
| Consensus MFE | -21.08 |
| Energy contribution | -20.00 |
| Covariance contribution | -1.08 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.889217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5205989 104 + 22422827 GCCUUCUUGGAUUGAAAAAAAA----GUGUUUUCAAAUGUCAGCGGCGCGGUGUGUUGAGGUGAGUUUGGCUUCGAACCUACCACGGCUAUGGCAACACUCGAGGAAA---------- ...(((((.(((((((((....----...))))))).(((((..(.((.((((.((((((((.......))))).))).)))).)).)..)))))....)).))))).---------- ( -29.20, z-score = -0.23, R) >droSim1.chrX_random 1800721 102 + 5698898 GCCUACUUGGAUCGAAAAAAA-----GUGUUUUCGAAUUUCAGUGCUGCGGUGAGUUGAGGUGAGUUUG-CUACGAACUCACCACAGCUGUGGCAACACUCAAGCAAA---------- ((..(((..(((((((((...-----...)))))).)))..)))))(((..(((((((.((((((((((-...)))))))))).)))).(((....)))))).)))..---------- ( -36.20, z-score = -2.84, R) >droSec1.super_4 4632377 102 - 6179234 GCCUUCUUGGAUCGAAAAAAA-----GUGUUUUCGAAUGUCAGUGCUGCGGUGAGUUGAGGUGAGUUUG-CCACGAACUCACCACAGCUAUGGCAACACUCAAGCAAA---------- .....(((((.(((((((...-----...))))))).(((((..((((.(((((((((.(((......)-)).).)))))))).))))..)))))....)))))....---------- ( -37.50, z-score = -3.18, R) >droYak2.chrX 18510684 117 - 21770863 GCUUACUUGGAUCGAAAAAAGUGUUCUUUUUUUCAAAAGCCAACUGUGCGGUGUGUUAAGGUGGGUACA-CUUGGUAUACACCAUGGCUGUGGCAGCACUGAAACAGGCGCGCUCAAA ......((((...((((((((....))))))))......))))..(((((((((((..(((((....))-)))...)))))))..(.((((..((....))..)))).)))))..... ( -36.90, z-score = -1.40, R) >droEre2.scaffold_4690 2560603 111 + 18748788 GCCUACUGGGAUUGGAAAAAAG------UGUUUCAAAUGACAGCGGUAUGGUGUGUUGGGGUGAGUAAG-CUUGGUAUUCAACAUGGCUGUGGCAACACUGAAACAGUCGCGCUCAAA (((..(((..(((.((((....------..)))).)))..))).))).((((((((((((.((((....-))))...))))))..((((((..((....))..)))))))))).)).. ( -27.70, z-score = 0.50, R) >consensus GCCUACUUGGAUCGAAAAAAA_____GUGUUUUCAAAUGUCAGCGGUGCGGUGUGUUGAGGUGAGUUUG_CUUCGAACUCACCACGGCUGUGGCAACACUCAAGCAAA__________ (((........(((((((...........)))))))..................((((.(((((((((......))))))))).))))...)))........................ (-21.08 = -20.00 + -1.08)

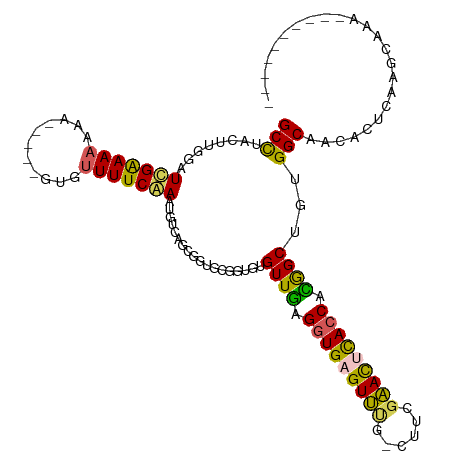
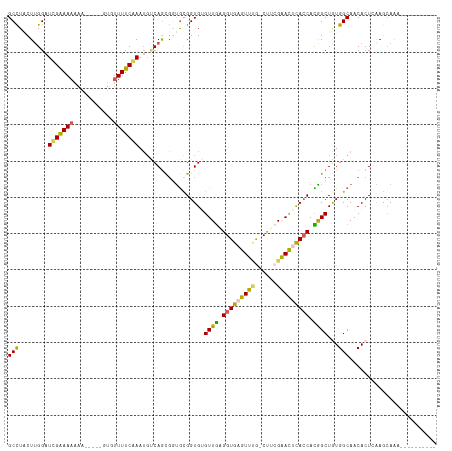
| Location | 5,205,989 – 5,206,093 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 71.05 |
| Shannon entropy | 0.47608 |
| G+C content | 0.46843 |
| Mean single sequence MFE | -27.74 |
| Consensus MFE | -13.87 |
| Energy contribution | -15.07 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.692452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5205989 104 - 22422827 ----------UUUCCUCGAGUGUUGCCAUAGCCGUGGUAGGUUCGAAGCCAAACUCACCUCAACACACCGCGCCGCUGACAUUUGAAAACAC----UUUUUUUUCAAUCCAAGAAGGC ----------.....(((((((((((....((.(((((((((..((........))))))..)).))).))...)).))))))))).....(----((((((........))))))). ( -21.10, z-score = 0.05, R) >droSim1.chrX_random 1800721 102 - 5698898 ----------UUUGCUUGAGUGUUGCCACAGCUGUGGUGAGUUCGUAG-CAAACUCACCUCAACUCACCGCAGCACUGAAAUUCGAAAACAC-----UUUUUUUCGAUCCAAGUAGGC ----------.(((((((((((((((...((.((.((((((((.....-..)))))))).)).))....)))))))).....(((((((...-----...)))))))..))))))).. ( -36.60, z-score = -4.11, R) >droSec1.super_4 4632377 102 + 6179234 ----------UUUGCUUGAGUGUUGCCAUAGCUGUGGUGAGUUCGUGG-CAAACUCACCUCAACUCACCGCAGCACUGACAUUCGAAAACAC-----UUUUUUUCGAUCCAAGAAGGC ----------..............(((...(((((((((((((.(.((-........)).)))))))))))))).((.....(((((((...-----...)))))))....))..))) ( -35.90, z-score = -3.51, R) >droYak2.chrX 18510684 117 + 21770863 UUUGAGCGCGCCUGUUUCAGUGCUGCCACAGCCAUGGUGUAUACCAAG-UGUACCCACCUUAACACACCGCACAGUUGGCUUUUGAAAAAAAGAACACUUUUUUCGAUCCAAGUAAGC .(((.(((.(..((((...(.((((...))))).(((.((((((...)-))))))))....))))..)))).)))((((...(((((((((.......))))))))).))))...... ( -25.90, z-score = -0.12, R) >droEre2.scaffold_4690 2560603 111 - 18748788 UUUGAGCGCGACUGUUUCAGUGUUGCCACAGCCAUGUUGAAUACCAAG-CUUACUCACCCCAACACACCAUACCGCUGUCAUUUGAAACA------CUUUUUUCCAAUCCCAGUAGGC ..((((.((......(((((..(.((....)).)..)))))......)-)...))))...............((((((..(((.((((..------....)))).)))..)))).)). ( -19.20, z-score = -0.27, R) >consensus __________UUUGCUUGAGUGUUGCCACAGCCGUGGUGAGUUCGAAG_CAAACUCACCUCAACACACCGCACCGCUGACAUUUGAAAACAC_____UUUUUUUCGAUCCAAGUAGGC ........................(((.((((((((((((((....................))))))))))..))))....(((((((...........)))))))........))) (-13.87 = -15.07 + 1.20)

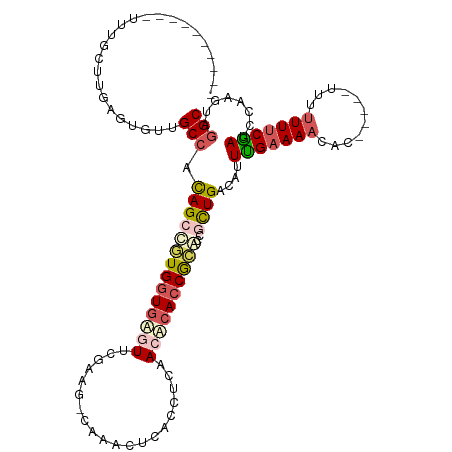
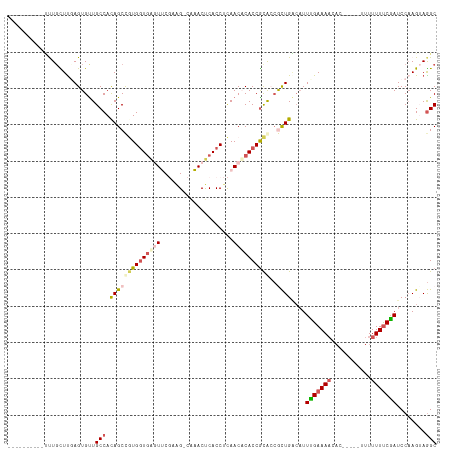
| Location | 5,206,014 – 5,206,118 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.63 |
| Shannon entropy | 0.53921 |
| G+C content | 0.47144 |
| Mean single sequence MFE | -35.22 |
| Consensus MFE | -19.51 |
| Energy contribution | -18.19 |
| Covariance contribution | -1.32 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.953853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5206014 104 + 22422827 -UUUUCAAAUGUCAGCGGCGCGGUGUGUUGAGGUGAGUUUGGCUUCGAACCUACCACGGCUAUGGCAACACUCGAGGA----------AAAUAUUAGUACU---AU--GUCGUGUGACGC -.........((((((((((..(((.((((.((((.(((((....))))).)))).))))..((....))........----------.........))).---.)--))))).)))).. ( -29.00, z-score = -0.09, R) >droSim1.chrX_random 1800745 106 + 5698898 -UUUUCGAAUUUCAGUGCUGCGGUGAGUUGAGGUGAGUUUG-CUACGAACUCACCACAGCUGUGGCAACACUCAAGCA----------AAAUAUUAGCAUUAGCAA--AUCGUGCGACGC -............(((((((.....(((((.((((((((((-...)))))))))).)))))(((....))).......----------......))))))).(((.--....)))..... ( -33.30, z-score = -1.62, R) >droSec1.super_4 4632401 106 - 6179234 -UUUUCGAAUGUCAGUGCUGCGGUGAGUUGAGGUGAGUUUG-CCACGAACUCACCACAGCUAUGGCAACACUCAAGCA----------AAAUAUUAGCAUUAGCAA--AUCGUGCGACGC -.........(((((((.(((.((.(((((.((((((((((-...)))))))))).))))))).))).))))...(((----------....(((.((....)).)--))..)))))).. ( -37.70, z-score = -3.05, R) >droYak2.chrX 18510712 116 - 21770863 UUUUUCAAAAGCCAACUGUGCGGUGUGUUAAGGUGGGUACA-CUUGGUAUACACCAUGGCUGUGGCAGCACUGAAACAGGCGCGCUCAAAAUAUUAGUGCCGCGAAUAGUGCUUUGA--- ....(((((.((((...((..(((((((..(((((....))-)))...)))))))...))..)))).((((((.....(((((.............))))).....)))))))))))--- ( -41.02, z-score = -1.78, R) >droEre2.scaffold_4690 2560627 109 + 18748788 --UUUCAAAUGACAGCGGUAUGGUGUGUUGGGGUGAGUAAG-CUUGGUAUUCAACAUGGCUGUGGCAACACUGAAACAGUCGCGCUCAAAAUGUUGGCGCUGUA-----UCAUUUGA--- --..(((((((((((((.((..((...(((((((((.(.((-((((........)).))))(((....)))......).)))).))))).))..)).)))))..-----))))))))--- ( -35.10, z-score = -1.76, R) >consensus _UUUUCAAAUGUCAGCGGUGCGGUGUGUUGAGGUGAGUUUG_CUUCGAACUCACCACGGCUGUGGCAACACUCAAGCA__________AAAUAUUAGCACUAGCAA__AUCGUGUGACGC ....(((.(((..........((((.((((.(((((((((......))))))))).)))).(((....)))..........................)))).........))).)))... (-19.51 = -18.19 + -1.32)

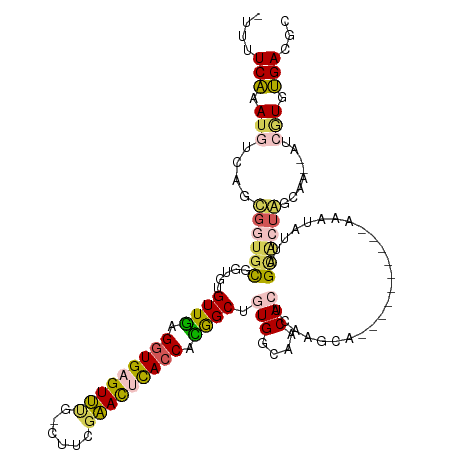
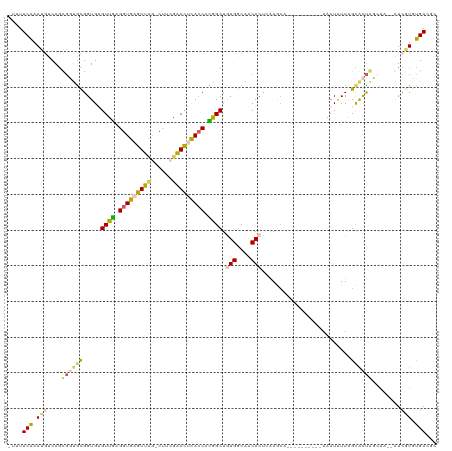
| Location | 5,206,014 – 5,206,118 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.63 |
| Shannon entropy | 0.53921 |
| G+C content | 0.47144 |
| Mean single sequence MFE | -28.93 |
| Consensus MFE | -13.19 |
| Energy contribution | -12.99 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.742824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5206014 104 - 22422827 GCGUCACACGAC--AU---AGUACUAAUAUUU----------UCCUCGAGUGUUGCCAUAGCCGUGGUAGGUUCGAAGCCAAACUCACCUCAACACACCGCGCCGCUGACAUUUGAAAA- ..(((....)))--..---.............----------...(((((((((((....((.(((((((((..((........))))))..)).))).))...)).)))))))))...- ( -21.80, z-score = -0.07, R) >droSim1.chrX_random 1800745 106 - 5698898 GCGUCGCACGAU--UUGCUAAUGCUAAUAUUU----------UGCUUGAGUGUUGCCACAGCUGUGGUGAGUUCGUAG-CAAACUCACCUCAACUCACCGCAGCACUGAAAUUCGAAAA- ...(((((.(((--..((....))....))).----------)))((.((((((((...((.((.((((((((.....-..)))))))).)).))....)))))))).))....))...- ( -31.70, z-score = -1.82, R) >droSec1.super_4 4632401 106 + 6179234 GCGUCGCACGAU--UUGCUAAUGCUAAUAUUU----------UGCUUGAGUGUUGCCAUAGCUGUGGUGAGUUCGUGG-CAAACUCACCUCAACUCACCGCAGCACUGACAUUCGAAAA- .....(((.(((--..((....))....))).----------)))(((((((((......(((((((((((((.(.((-........)).))))))))))))))...)))))))))...- ( -37.80, z-score = -3.41, R) >droYak2.chrX 18510712 116 + 21770863 ---UCAAAGCACUAUUCGCGGCACUAAUAUUUUGAGCGCGCCUGUUUCAGUGCUGCCACAGCCAUGGUGUAUACCAAG-UGUACCCACCUUAACACACCGCACAGUUGGCUUUUGAAAAA ---(((((((.......((((((((........(((((....))))).))))))))..((((..((((((.......(-((....)))......))))))....)))))).))))).... ( -27.02, z-score = 0.36, R) >droEre2.scaffold_4690 2560627 109 - 18748788 ---UCAAAUGA-----UACAGCGCCAACAUUUUGAGCGCGACUGUUUCAGUGUUGCCACAGCCAUGUUGAAUACCAAG-CUUACUCACCCCAACACACCAUACCGCUGUCAUUUGAAA-- ---((((((((-----(((.(((((((....))).))))..........((((((..........(((........))-)..........))))))........).))))))))))..-- ( -26.35, z-score = -2.68, R) >consensus GCGUCACACGAC__UUGCCAGUGCUAAUAUUU__________UGCUUGAGUGUUGCCACAGCCGUGGUGAGUUCGAAG_CAAACUCACCUCAACACACCGCACCGCUGACAUUUGAAAA_ ...(((............(((((((.......................)))))))...((((((((((((((....................))))))))))..)))).....))).... (-13.19 = -12.99 + -0.20)

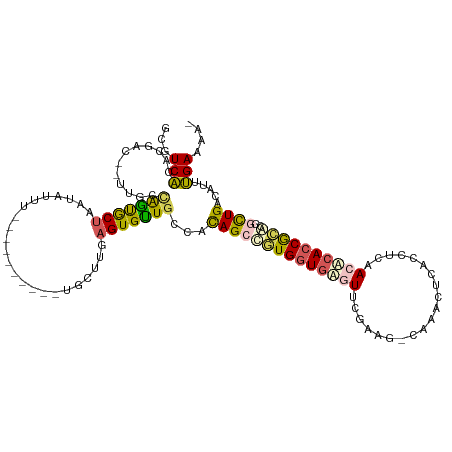
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:17:15 2011