
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,128,031 – 5,128,143 |
| Length | 112 |
| Max. P | 0.700362 |

| Location | 5,128,031 – 5,128,143 |
|---|---|
| Length | 112 |
| Sequences | 14 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 76.36 |
| Shannon entropy | 0.54032 |
| G+C content | 0.50795 |
| Mean single sequence MFE | -31.61 |
| Consensus MFE | -16.38 |
| Energy contribution | -16.88 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.700362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
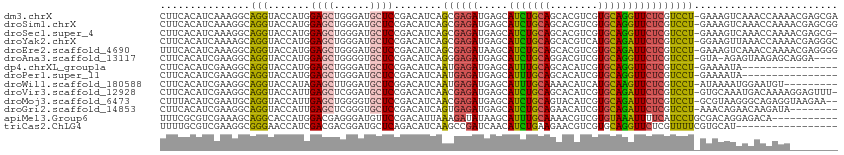
>dm3.chrX 5128031 112 + 22422827 UCGCUCGUUUUGGUUUGACUUUC-AGGACGAGAACCUGCACGACGUGCUGCAGAUGCUCAUCUCGCUGAUGUCGGAGCAUCCCAGCUCCAUGGUACCUGCCUUUGAUGUGAAG ...((((((((((........))-))))))))......((((((((.(.((((((....)))).)).))))))(((((......)))))..(((....)))......)))... ( -35.90, z-score = -0.90, R) >droSim1.chrX 3961052 112 + 17042790 CCGCUCGUUUUGGUUUGACUUUC-AGGACGAGAACCUGCACGACGUGCUGCAGAUGCUCAUCUCGCUGAUGUCGGAGCAUCCCAGCUCCAUGGUACCUGCCUUUGAUGUGAAG ...((((((((((........))-))))))))......((((((((.(.((((((....)))).)).))))))(((((......)))))..(((....)))......)))... ( -35.90, z-score = -1.03, R) >droSec1.super_4 4550116 111 - 6179234 -CGCUCGUUUUGGUUUGACUUUC-AGGACGAGAACCUGCACGACGUGCUGCAGAUGCUCAUCUCGCUGAUGUCGGAGCAUCCCAGCUCCAUGGUACCUGCCUUUGAUGUGAAG -..((((((((((........))-))))))))......((((((((.(.((((((....)))).)).))))))(((((......)))))..(((....)))......)))... ( -35.90, z-score = -1.01, R) >droYak2.chrX 18433121 112 - 21770863 GCCCUCGUUUUGGUUUAACUUCC-AGGACGAGAAUCUGCAUGACGUGCUGCAGAUGCUCAUCUCGCUGAUGUCGGAGCAUCCCAGCUCCAUGGUACCUGCUUUUGAUGUGAAG ...((((((((((........))-)))))))).(((.((((...)))).((((.((((((((.....))))..(((((......)))))..)))).))))....)))...... ( -37.20, z-score = -1.89, R) >droEre2.scaffold_4690 2482912 112 + 18748788 CCCCUCGUUUUGGUUUGACUUUC-AGGACGAGAAUCUGCACGACGUGCUGCAGAUGCUUAUCUCGCUGAUGUCGGAGCAUCCCAGCUCCAUGGUACCUGCCUUUGAUGUGAAA ...((((((((((........))-))))))))......((((((((.(.((((((....)))).)).))))))(((((......)))))..(((....)))......)))... ( -35.00, z-score = -1.60, R) >droAna3.scaffold_13117 2558508 107 + 5790199 ----UCCUGCUCUUACUCU-UAC-AGGACGAGAACCUGCACGACGUCCUGCAGAUGCUCAUCUCCCUGAUGUCGGAGCACCCCAGCUCCAUGGUACCUGCCUUCGAUGUGAAG ----(((((..........-..)-))))..........((((.((....((((.((((((((.....))))..(((((......)))))..)))).))))...)).))))... ( -29.90, z-score = -1.10, R) >dp4.chrXL_group1a 5122163 96 - 9151740 ----------------UAUUUUC-AGGACGAGAACCUGCACGAUGUGCUGCAAAUGCUCAUCUCAUUGAUGUCGGAGCAUCCCAGCUCCAUGGUACCUGCCUUCGAUGUGAAG ----------------......(-(((.......))))(((((((.((.......)).))))..(((((....(((((......)))))..(((....))).))))))))... ( -26.40, z-score = -0.66, R) >droPer1.super_11 2250338 96 - 2846995 ----------------UAUUUUC-AGGACGAGAACCUGCACGAUGUGCUGCAAAUGCUCAUCUCAUUGAUGUCGGAGCAUCCCAGCUCCAUGGUACCUGCCUUCGAUGUGAAG ----------------......(-(((.......))))(((((((.((.......)).))))..(((((....(((((......)))))..(((....))).))))))))... ( -26.40, z-score = -0.66, R) >droWil1.scaffold_180588 819215 103 - 1294757 ---------ACAUUCCAUUUUAU-AGGACGAGAACUUGCAUGAUGUUUUGCAAAUGCUCAUCUCAUUGAUGUCCGAGCAUCCAAGCUCUAUGGUACCUGCCUUCGAUGUGAAG ---------...(((((((....-.(((((((...(((((........)))))...)))(((.....)))))))((((......))))...(((....)))...)))).))). ( -26.40, z-score = -1.49, R) >droVir3.scaffold_12928 3165678 111 + 7717345 -AAACUCCUUUUGUCAUUUGCAC-AGGACGAGAAUCUGCACGAUGUGCUGCAGAUGCUCAUCUCGUUGAUGUCGGAGCAUCCGAGCUCAAUGGUACCUGCCUUCGAUGUGAAG -......((((...((((....(-((((((((.(((((((........))))))).)))....((((((..((((.....))))..)))))))).)))).....)))).)))) ( -33.90, z-score = -0.92, R) >droMoj3.scaffold_6473 16571251 110 - 16943266 --UUCUUACCUCUGCCCUUACGC-AGGACGAGAAUCUGCACGAUGUACUGCAGAUGCUCAUCUCGUUGAUGUCGGAGCACCCCAGCUCAAUGGUACCUGCAUUCGAUGUAAAG --...((((.((.(......)((-((((((((.(((((((........))))))).)))....((((((.((.((......)).)))))))))).)))))....)).)))).. ( -33.10, z-score = -1.92, R) >droGri2.scaffold_14853 2286762 104 + 10151454 --------UAUCUUGUUCUGUUU-AGGACGAGAAUCUGCACGAUGUUCUGCAGAUGCUCAUCUCACUGAUGUCGGAGCACCCGAGCUCAAUGGUACCUGCCUUCGAUGUGAAG --------..(((((((((....-)))))))))(((((((.(.....)))))))).......((((....((((((((......))))...(((....)))..)))))))).. ( -31.60, z-score = -1.26, R) >apiMel3.Group6 8813824 102 + 14581788 -----------UGUCUCCUGUCGCAGGAUGAAAAUUUACACGACGUUUUGCAAAUGCUUAUAUCUUUAAUGUCGGAACAUCCCUCGUCCAUGGUGCCUGCUUUCGACGCGAAA -----------.........((((.((((((.........(((((((..((....))..........))))))).........))))))..(((....)))......)))).. ( -20.97, z-score = 0.11, R) >triCas2.ChLG4 10636591 96 - 13894384 -----------------AUGCACGAAAACGAGAACCUGCACGACGUUCUUCAGAUGUUGAUCGGCUUGAUGUCUGAGCAUCCGUCGUCGAUGGUUCCCGCCUUCGACGCAAAA -----------------.(((.((((..((.(((((...((((((...(((((((((..(.....)..)))))))))....))))))....))))).))..))))..)))... ( -34.00, z-score = -2.40, R) >consensus ________UUUGGUUUUAUUUUC_AGGACGAGAACCUGCACGACGUGCUGCAGAUGCUCAUCUCGCUGAUGUCGGAGCAUCCCAGCUCCAUGGUACCUGCCUUCGAUGUGAAG .........................(((.(((...(((((........)))))...))).))).....((((((((((......))))...(((....)))..)))))).... (-16.38 = -16.88 + 0.50)



| Location | 5,128,031 – 5,128,143 |
|---|---|
| Length | 112 |
| Sequences | 14 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 76.36 |
| Shannon entropy | 0.54032 |
| G+C content | 0.50795 |
| Mean single sequence MFE | -33.34 |
| Consensus MFE | -16.32 |
| Energy contribution | -15.88 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.644754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
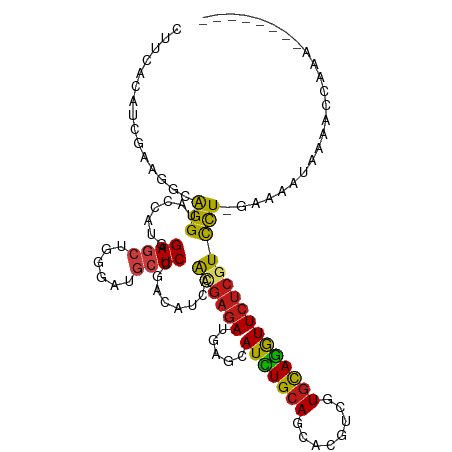
>dm3.chrX 5128031 112 - 22422827 CUUCACAUCAAAGGCAGGUACCAUGGAGCUGGGAUGCUCCGACAUCAGCGAGAUGAGCAUCUGCAGCACGUCGUGCAGGUUCUCGUCCU-GAAAGUCAAACCAAAACGAGCGA ................(((.....(((((......)))))(((.((((...((((((.(((((((........))))))).))))))))-))..)))..)))........... ( -35.40, z-score = -0.76, R) >droSim1.chrX 3961052 112 - 17042790 CUUCACAUCAAAGGCAGGUACCAUGGAGCUGGGAUGCUCCGACAUCAGCGAGAUGAGCAUCUGCAGCACGUCGUGCAGGUUCUCGUCCU-GAAAGUCAAACCAAAACGAGCGG ................(((.....(((((......)))))(((.((((...((((((.(((((((........))))))).))))))))-))..)))..)))........... ( -35.40, z-score = -0.82, R) >droSec1.super_4 4550116 111 + 6179234 CUUCACAUCAAAGGCAGGUACCAUGGAGCUGGGAUGCUCCGACAUCAGCGAGAUGAGCAUCUGCAGCACGUCGUGCAGGUUCUCGUCCU-GAAAGUCAAACCAAAACGAGCG- ................(((.....(((((......)))))(((.((((...((((((.(((((((........))))))).))))))))-))..)))..)))..........- ( -35.40, z-score = -1.02, R) >droYak2.chrX 18433121 112 + 21770863 CUUCACAUCAAAAGCAGGUACCAUGGAGCUGGGAUGCUCCGACAUCAGCGAGAUGAGCAUCUGCAGCACGUCAUGCAGAUUCUCGUCCU-GGAAGUUAAACCAAAACGAGGGC ((((.........((.(((....((((((......))))))..))).))..((((((.(((((((........))))))).)))))).(-((........)))....)))).. ( -34.90, z-score = -1.31, R) >droEre2.scaffold_4690 2482912 112 - 18748788 UUUCACAUCAAAGGCAGGUACCAUGGAGCUGGGAUGCUCCGACAUCAGCGAGAUAAGCAUCUGCAGCACGUCGUGCAGAUUCUCGUCCU-GAAAGUCAAACCAAAACGAGGGG ................(((.....(((((......)))))(((.(((((((((.....(((((((........))))))))))))..))-))..)))..)))........... ( -32.10, z-score = -0.73, R) >droAna3.scaffold_13117 2558508 107 - 5790199 CUUCACAUCGAAGGCAGGUACCAUGGAGCUGGGGUGCUCCGACAUCAGGGAGAUGAGCAUCUGCAGGACGUCGUGCAGGUUCUCGUCCU-GUA-AGAGUAAGAGCAGGA---- ........(((..((..((((...(((((......)))))(((.((..(.((((....)))).)..)).)))))))..))..)))((((-((.-.........))))))---- ( -37.50, z-score = -0.82, R) >dp4.chrXL_group1a 5122163 96 + 9151740 CUUCACAUCGAAGGCAGGUACCAUGGAGCUGGGAUGCUCCGACAUCAAUGAGAUGAGCAUUUGCAGCACAUCGUGCAGGUUCUCGUCCU-GAAAAUA---------------- .((((...(((..((..((((.(((..((((((((((((....(((.....))))))))))).)))).))).))))..))..)))...)-)))....---------------- ( -32.50, z-score = -1.78, R) >droPer1.super_11 2250338 96 + 2846995 CUUCACAUCGAAGGCAGGUACCAUGGAGCUGGGAUGCUCCGACAUCAAUGAGAUGAGCAUUUGCAGCACAUCGUGCAGGUUCUCGUCCU-GAAAAUA---------------- .((((...(((..((..((((.(((..((((((((((((....(((.....))))))))))).)))).))).))))..))..)))...)-)))....---------------- ( -32.50, z-score = -1.78, R) >droWil1.scaffold_180588 819215 103 + 1294757 CUUCACAUCGAAGGCAGGUACCAUAGAGCUUGGAUGCUCGGACAUCAAUGAGAUGAGCAUUUGCAAAACAUCAUGCAAGUUCUCGUCCU-AUAAAAUGGAAUGU--------- ((((.....))))(((....((((.((((......))))(((((((.....)))(((.(((((((........))))))).))))))).-.....))))..)))--------- ( -30.30, z-score = -2.26, R) >droVir3.scaffold_12928 3165678 111 - 7717345 CUUCACAUCGAAGGCAGGUACCAUUGAGCUCGGAUGCUCCGACAUCAACGAGAUGAGCAUCUGCAGCACAUCGUGCAGAUUCUCGUCCU-GUGCAAAUGACAAAAGGAGUUU- ((((.....))))((((......((((..((((.....))))..))))...((((((.(((((((........))))))).))))))))-))....................- ( -32.40, z-score = -0.67, R) >droMoj3.scaffold_6473 16571251 110 + 16943266 CUUUACAUCGAAUGCAGGUACCAUUGAGCUGGGGUGCUCCGACAUCAACGAGAUGAGCAUCUGCAGUACAUCGUGCAGAUUCUCGUCCU-GCGUAAGGGCAGAGGUAAGAA-- .(((((.((((((....((((...((.((((((((((((....(((.....))))))))))).)))).))..))))..))))..(((((-.....))))).)).)))))..-- ( -33.60, z-score = -0.55, R) >droGri2.scaffold_14853 2286762 104 - 10151454 CUUCACAUCGAAGGCAGGUACCAUUGAGCUCGGGUGCUCCGACAUCAGUGAGAUGAGCAUCUGCAGAACAUCGUGCAGAUUCUCGUCCU-AAACAGAACAAGAUA-------- ((((.....))))........((((((..((((.....))))..)))))).((((((.((((((((.....).))))))).))))))..-...............-------- ( -31.70, z-score = -1.51, R) >apiMel3.Group6 8813824 102 - 14581788 UUUCGCGUCGAAAGCAGGCACCAUGGACGAGGGAUGUUCCGACAUUAAAGAUAUAAGCAUUUGCAAAACGUCGUGUAAAUUUUCAUCCUGCGACAGGAGACA----------- ......((((((((...((((....((((...(((((....)))))..........((....))....))))))))...))))).(((((...)))))))).----------- ( -24.10, z-score = -0.36, R) >triCas2.ChLG4 10636591 96 + 13894384 UUUUGCGUCGAAGGCGGGAACCAUCGACGACGGAUGCUCAGACAUCAAGCCGAUCAACAUCUGAAGAACGUCGUGCAGGUUCUCGUUUUCGUGCAU----------------- ...((((.((((((((((((((....((((((..(..(((((.(((.....))).....))))).)..))))))...)))))))))))))))))).----------------- ( -39.00, z-score = -3.30, R) >consensus CUUCACAUCGAAGGCAGGUACCAUGGAGCUGGGAUGCUCCGACAUCAACGAGAUGAGCAUCUGCAGCACGUCGUGCAGGUUCUCGUCCU_GAAAAUAAAACCAAA________ ...............(((.......((((......))))........((((((.....(((((((........))))))))))))))))........................ (-16.32 = -15.88 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:17:08 2011