| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,081,620 – 5,081,696 |
| Length | 76 |
| Max. P | 0.684809 |

| Location | 5,081,620 – 5,081,696 |
|---|---|
| Length | 76 |
| Sequences | 11 |
| Columns | 85 |
| Reading direction | forward |
| Mean pairwise identity | 70.36 |
| Shannon entropy | 0.58643 |
| G+C content | 0.38285 |
| Mean single sequence MFE | -7.79 |
| Consensus MFE | -5.70 |
| Energy contribution | -5.79 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.27 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.684809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
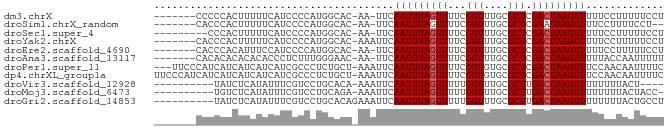
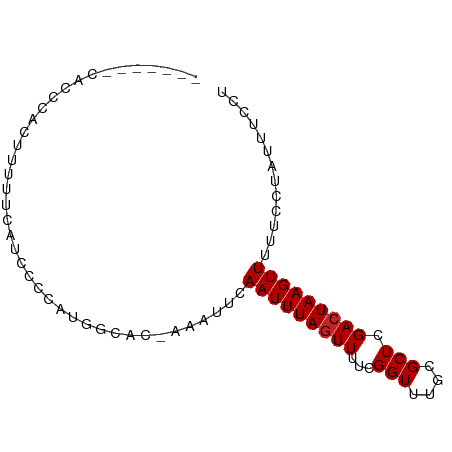
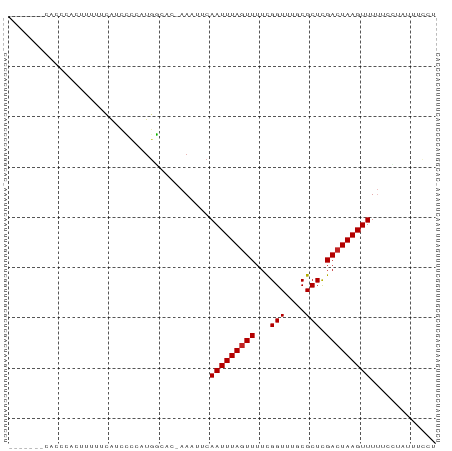
>dm3.chrX 5081620 76 + 22422827 -------CCCCCACUUUUUCAUCCCCAUGGCAC-AA-UUCAAUUUAGUUUUCGGUUUGCGCUCGACUAAGUUUUUCCUUUUUCCU -------..........................-..-...(((((((((..((.....))...)))))))))............. ( -5.80, z-score = 0.43, R) >droSim1.chrX_random 1763049 74 + 5698898 -------CACCCACUUUUUCAUCCCCAUGGCAC-AA-UUCAAUUUAGUUUUCGGUUUGCGCUCGAAUAAGUUUUCCUUUUCCU-- -------.....((((.(((........(((.(-((-..(............)..))).))).))).))))............-- ( -4.40, z-score = 1.13, R) >droSec1.super_4 4505495 74 - 6179234 ---------CCCACUUUUUCAUCCCCAUGGCAC-AA-UUCAAUUUAGUUUUCGGUUUGCGCUCGACUAAGUUUUUCCUUUUUCCU ---------........................-..-...(((((((((..((.....))...)))))))))............. ( -5.80, z-score = 0.33, R) >droYak2.chrX 18385097 77 - 21770863 -------CACCCACUUUUUCAUCCCCAUGGCAC-AAAUUCAAUUUAGUUUUCGGUUUGCGCUCGACUAAGUUUUUCCUUUUUCCU -------.....................(((.(-((((..((.......))..))))).))).((..(((......)))..)).. ( -6.60, z-score = 0.04, R) >droEre2.scaffold_4690 2436911 76 + 18748788 -------CACCCACAUUUCCAUCCCCAUGGCAC-AA-UUCAAUUUAGUUUUCGGUUUGCGCUCGACUAAGUUUUUCCUUUUUCCU -------...........((((....))))...-..-...(((((((((..((.....))...)))))))))............. ( -7.30, z-score = -0.31, R) >droAna3.scaffold_13117 2513228 76 + 5790199 -------CACACACACACACCCUCUUUGGGAAC-AA-UUCAAUUUAGUUUUCGGUUUGCGCUCGACUAAGUUUUUACCAAUUUUU -------............(((.....)))...-..-...(((((((((..((.....))...)))))))))............. ( -10.40, z-score = -1.11, R) >droPer1.super_11 2201408 81 - 2846995 ---UUCCCAUCAUCAUCAUCAUCGCCCUCUGCU-AAAUUCAAUUUAGUUUUCGGUGUGCGCUCGACUAAGUUUCCAACAAUUUUC ---................((.((((....(((-((((...)))))))....))))))........................... ( -8.50, z-score = -0.74, R) >dp4.chrXL_group1a 5072988 84 - 9151740 UUCCCAUCAUCAUCAUCAUCAUCGCCCUCUGCU-AAAUUCAAUUUAGUUUUCGGUGUGCGCUCGACUAAGUUUCCAACAAUUUUC ...................((.((((....(((-((((...)))))))....))))))........................... ( -8.50, z-score = -0.85, R) >droVir3.scaffold_12928 3087542 70 + 7717345 ----------UAUCUCAUAUUUCGUCCUGCACA-AAAUUCAAUUUAGUUUUUGGUUUGCGCUUGACUAAGUUUUUUUUACU---- ----------........((((.(((..((.((-((..((((........)))))))).))..))).))))..........---- ( -9.60, z-score = -1.40, R) >droMoj3.scaffold_6473 16494287 73 - 16943266 ----------UGUCUCAUAUUUCGUCCUGCAGA-AAAUUCAAUUUAGUUUUUGGUUUGCGCUUGACUAAGUUUUUUUUACUACC- ----------.........((((........))-))....(((((((((...(((....))).)))))))))............- ( -8.40, z-score = 0.05, R) >droGri2.scaffold_14853 2222463 75 + 10151454 ----------UAUCUCAUAUUUCGUCCUGCACAGAAAUUCAAUUUAGUUUUUGGUUUGCGCUUGACUAAGUUUUUUUUACUGCCU ----------..................(((..((((...(((((((((...(((....))).)))))))))..))))..))).. ( -10.40, z-score = -0.60, R) >consensus _______CACCCACUUUUUCAUCCCCAUGGCAC_AAAUUCAAUUUAGUUUUCGGUUUGCGCUCGACUAAGUUUUUCCUAUUUCCU ........................................(((((((((...(((....))).)))))))))............. ( -5.70 = -5.79 + 0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:16:58 2011