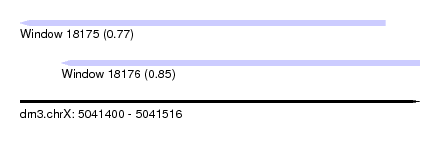
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,041,400 – 5,041,516 |
| Length | 116 |
| Max. P | 0.850635 |

| Location | 5,041,400 – 5,041,506 |
|---|---|
| Length | 106 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 67.85 |
| Shannon entropy | 0.62283 |
| G+C content | 0.60119 |
| Mean single sequence MFE | -32.51 |
| Consensus MFE | -10.73 |
| Energy contribution | -11.19 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.773058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
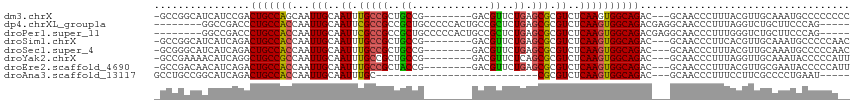
>dm3.chrX 5041400 106 - 22422827 UCCGACUGCCAGCAAUUGCAAUUUGCCGCUGCCG--------GACGUUCUGAGCGCGUCUCAAGUGGCAGAC---GCAACCCUUUACGUUGCAAAUGCCCCCCCCCCAACACCCCCC ...........(((.......(((((((((...(--------(((((.......))))))..))))))))).---(((((.......)))))...)))................... ( -28.30, z-score = -1.99, R) >dp4.chrXL_group1a 5023920 107 + 9151740 -----CUGCCACCAAUUGCAAUUCGCCGCCGCUGCCCCCACUGCCGCUCUGAGCGCGUCUCAAGUGGCAGACGAGGCAACCCUUUAGGUC-----UGCUUCCCAGCUCCUGCCUGCU -----............(((....((.(..((((......((((((((.((((.....))))))))))))..((((((..((....))..-----)))))).))))..).)).))). ( -35.70, z-score = -1.36, R) >droPer1.super_11 2155202 107 + 2846995 -----CUGCCACCAAUUGCAAUUCGCCGCCGCUGCCCCCACUGCCGCUCUGAGCGCGUCUCAAGUGGCAGACGAGGCAACCCUUUGGGUC-----UGCUUCCCAGCUGCUGCCUGCU -----............(((....((.((.((((......((((((((.((((.....))))))))))))..((((((((((...)))).-----)))))).)))).)).)).))). ( -41.40, z-score = -1.95, R) >droSim1.chrX 3889462 106 - 17042790 UCAGACUGCCACCAAUUGCAAUUUGCCGCUGCCG--------GACGUUCUGAGCGCGUCUCAAGUGGCAGAC---GCAACCCUUCACGUUGCAAAUGCCCCCAACACACAACCCCCU .................(((.(((((((((...(--------(((((.......))))))..))))))))).---(((((.......)))))...)))................... ( -28.10, z-score = -1.57, R) >droSec1.super_4 4466320 106 + 6179234 UCAGACUGCCACCAAUUGCAAUUUGCCGCUGCCG--------GACGUUCUGAGCGCGUCUCAAGUGGCAGAC---GCAACCCUUUACGUUGCAAAUGCCCCCAACACACAGCCCCCU .....(((.........(((.(((((((((...(--------(((((.......))))))..))))))))).---(((((.......)))))...)))..........)))...... ( -28.71, z-score = -1.43, R) >droYak2.chrX 18344029 101 + 21770863 UCAGGCUGCCGCCAAUUGCAAUUUGCCGCUGCCG--------GACGUUCUCAGCGCGUCUCAAGUGGCAGAC---GCAACCCUUUAGGUUGCAAAUACCCCCAUUACCCCCC----- ...(((....)))........(((((((((...(--------(((((.......))))))..))))))))).---((((((.....))))))....................----- ( -33.90, z-score = -2.81, R) >droEre2.scaffold_4690 2397486 94 - 18748788 UCAGACUGCCACCAAUUGCAAUUUGCCGCUACCG--------GACGUUCUGAGCGCGUCUCAAGUGGCAGAC---GCAACCCUUUACGUUGCGAAUACCCCCAUU------------ ......(((........))).(((((((((...(--------(((((.......))))))..)))))))))(---(((((.......))))))............------------ ( -27.80, z-score = -2.50, R) >droMoj3.scaffold_6473 16421574 94 + 16943266 -------GCCGGCUUGUGGUGCUUGCUGCUGCGGC-----UAGCCGUUCGGCCGGCUCUUUAAAGAGC-GAC--GGCUGCAUCCCAAGC-----AUCCCACUUCCACACCUCCC--- -------...((...((((((((((..(.((((((-----(.(((....)))..(((((....)))))-...--))))))).).)))))-----)..))))..)).........--- ( -36.20, z-score = -1.51, R) >consensus UCAGACUGCCACCAAUUGCAAUUUGCCGCUGCCG________GACGUUCUGAGCGCGUCUCAAGUGGCAGAC___GCAACCCUUUACGUUGCAAAUGCCCCCAACACACCGCCC_CU .....(((((((.....((.....((....))..........(.(((.....)))))).....)))))))............................................... (-10.73 = -11.19 + 0.45)
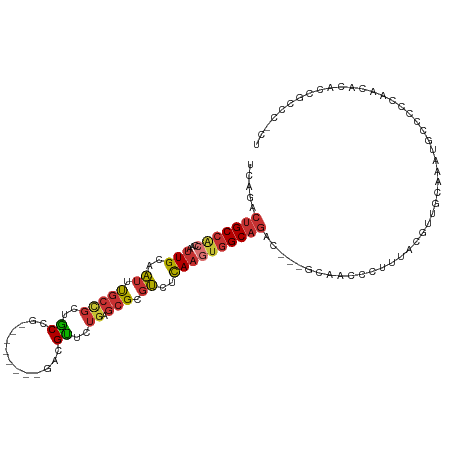
| Location | 5,041,412 – 5,041,516 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 71.32 |
| Shannon entropy | 0.55136 |
| G+C content | 0.59574 |
| Mean single sequence MFE | -33.08 |
| Consensus MFE | -13.07 |
| Energy contribution | -13.21 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.850635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 5041412 104 - 22422827 -GCCGGCAUCAUCCGACUGCCAGCAAUUGCAAUUUGCCGCUGCCG--------GACGUUCUGAGCGCGUCUCAAGUGGCAGAC---GCAACCCUUUACGUUGCAAAUGCCCCCCCC -((.((((((....)).)))).))....(((.(((((((((...(--------(((((.......))))))..))))))))).---(((((.......)))))...)))....... ( -37.20, z-score = -2.79, R) >dp4.chrXL_group1a 5023932 103 + 9151740 --------GGCCGACCCUGCCACCAAUUGCAAUUCGCCGCCGCUGCCCCCACUGCCGCUCUGAGCGCGUCUCAAGUGGCAGACGAGGCAACCCUUUAGGUCUGCUUCCCAG----- --------((((((...(((........)))..)))..))).(((......((((((((.((((.....))))))))))))..((((((..((....))..)))))).)))----- ( -35.40, z-score = -1.73, R) >droPer1.super_11 2155214 103 + 2846995 --------GGCCGACCCUGCCACCAAUUGCAAUUCGCCGCCGCUGCCCCCACUGCCGCUCUGAGCGCGUCUCAAGUGGCAGACGAGGCAACCCUUUGGGUCUGCUUCCCAG----- --------((((((...(((........)))..)))..))).(((......((((((((.((((.....))))))))))))..((((((((((...)))).)))))).)))----- ( -36.90, z-score = -1.46, R) >droSim1.chrX 3889474 104 - 17042790 -GCCGGCAUCAUCAGACUGCCACCAAUUGCAAUUUGCCGCUGCCG--------GACGUUCUGAGCGCGUCUCAAGUGGCAGAC---GCAACCCUUCACGUUGCAAAUGCCCCCAAC -...((((((....)).)))).......(((.(((((((((...(--------(((((.......))))))..))))))))).---(((((.......)))))...)))....... ( -34.50, z-score = -2.02, R) >droSec1.super_4 4466332 104 + 6179234 -GCGGGCAUCAUCAGACUGCCACCAAUUGCAAUUUGCCGCUGCCG--------GACGUUCUGAGCGCGUCUCAAGUGGCAGAC---GCAACCCUUUACGUUGCAAAUGCCCCCAAC -(.((((((.......(((((((.....((.....))(((.((((--------(.....))).)))))......)))))))..---(((((.......)))))..)))))).)... ( -37.10, z-score = -2.37, R) >droYak2.chrX 18344036 104 + 21770863 -GCCGAAAACAUCAGGCUGCCGCCAAUUGCAAUUUGCCGCUGCCG--------GACGUUCUCAGCGCGUCUCAAGUGGCAGAC---GCAACCCUUUAGGUUGCAAAUACCCCCAUU -((.((.....)).(((....)))....))..(((((((((...(--------(((((.......))))))..))))))))).---((((((.....))))))............. ( -34.30, z-score = -1.99, R) >droEre2.scaffold_4690 2397486 104 - 18748788 -GCCGACAACAUCAGACUGCCACCAAUUGCAAUUUGCCGCUACCG--------GACGUUCUGAGCGCGUCUCAAGUGGCAGAC---GCAACCCUUUACGUUGCGAAUACCCCCAUU -................(((........))).(((((((((...(--------(((((.......))))))..)))))))))(---(((((.......))))))............ ( -27.80, z-score = -1.92, R) >droAna3.scaffold_13117 2475974 81 - 5790199 GCCUGCCGGCAUCAGACUGCCACCAAUUGCAAUUUGC---------------------------CGCGUCUCAAGUGGCAGAC---GCAACCCUUUCCUUCGCCCCUGAAU----- (((....))).((((.(((((((....(((.......---------------------------.)))......)))))))..---((.............))..))))..----- ( -21.42, z-score = -1.25, R) >consensus _GCCGGCAGCAUCAGACUGCCACCAAUUGCAAUUUGCCGCUGCCG________GACGUUCUGAGCGCGUCUCAAGUGGCAGAC___GCAACCCUUUACGUUGCAAAUGCCCCCA__ ................(((((((.....((.....))..................(((.....)))........)))))))................................... (-13.07 = -13.21 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:16:56 2011