
| Sequence ID | dm3.chrX |
|---|---|
| Location | 5,039,815 – 5,039,887 |
| Length | 72 |
| Max. P | 0.650537 |

| Location | 5,039,815 – 5,039,887 |
|---|---|
| Length | 72 |
| Sequences | 8 |
| Columns | 79 |
| Reading direction | reverse |
| Mean pairwise identity | 74.86 |
| Shannon entropy | 0.50767 |
| G+C content | 0.41242 |
| Mean single sequence MFE | -14.06 |
| Consensus MFE | -6.65 |
| Energy contribution | -7.15 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.650537 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 5039815 72 - 22422827 GUAAAUACAAUUGCAAUUUCC-UGGGCUCUAUUUC----UGUGUGCACUUUUCACUUAAUUGCACAC--AGUUACGCAC ((((......)))).......-...((..((...(----((((((((.((.......)).)))))))--)).)).)).. ( -15.10, z-score = -2.07, R) >droAna3.scaffold_13117 2474551 55 - 5790199 GCAAAUACAAUUGCAAUUUCC--------------------UGUGCACUUUUCACUUAAUUGCACCC--ACACAGCC-- ((((......)))).......--------------------.(((((.((.......)).)))))..--........-- ( -7.70, z-score = -1.40, R) >droEre2.scaffold_4690 2395904 72 - 18748788 GCAAAUACAAUUGCAAUUUCC-UGGGCUCUAUUUC----UGUGUGCACUUUUCACUUAAUUGCACAC--UGGUACGCAC ((((......)))).......-...((..((((..----.(((((((.((.......)).)))))))--.)))).)).. ( -14.80, z-score = -1.54, R) >droSim1.chrX 3887543 72 - 17042790 GCAAAUACAAUUGCAAUUUCC-UGGGCUCUAUUUC----UGUGUGCACUUUUCACUUAAUUGCACAC--AGUUACGCAC ((((......)))).......-...((..((...(----((((((((.((.......)).)))))))--)).)).)).. ( -17.10, z-score = -2.73, R) >droWil1.scaffold_180588 725041 77 + 1294757 ACAAAUACAAAUGAAAUUUCCCGUUACAUCAUUGCAGUUUGUGUGCACUUUUCACUUAAUUGCACAC--AUUUACGCAC .........((((........)))).......(((.((.((((((((.((.......)).)))))))--)...))))). ( -14.20, z-score = -1.86, R) >droGri2.scaffold_14853 2164030 72 - 10151454 GCAAAUACAAUUGCAUUUUCC-GCCACUUCAUUGC----UAUGUGCACUUUUCACUUAAUUGCUCAC--AUUCACACAC ((((........((.......-))........(((----.....)))............))))....--.......... ( -5.50, z-score = 0.34, R) >dp4.chrXL_group1a 5019505 75 + 9151740 ACACACACAAUUGCACUUUCCCGGCUUCCCGUGUG----UGUGUGCACUUUUCAUUUAAAUGCACACGGAAUUACGCAC ..((((((....((.(......))).....)))))----)(((((((.(((......))))))))))............ ( -18.30, z-score = -1.45, R) >droPer1.super_11 2150882 75 + 2846995 ACACACACAAUUGCACUUUCCCGGCUGCUCGUGUG----UGUGUGCACUUUUCAUUUAAAUGCACACGGAAUUACGCAC ..((((((....(((((.....)).)))..)))))----)(((((((.(((......))))))))))............ ( -19.80, z-score = -1.43, R) >consensus GCAAAUACAAUUGCAAUUUCC_GGCGCUCCAUUUC____UGUGUGCACUUUUCACUUAAUUGCACAC__AGUUACGCAC ........................................(((((((.((.......)).)))))))............ ( -6.65 = -7.15 + 0.50)
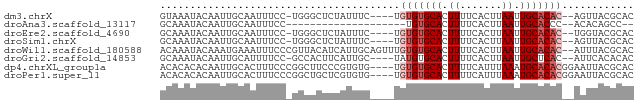

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:16:54 2011